Search Count: 272
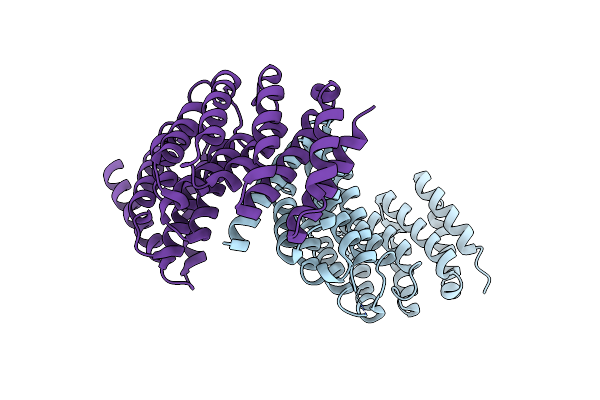 |
Crystal Structure Of Spiral2 Microtubule-Binding Domain From Physcomitrella Patens
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: PLANT PROTEIN |
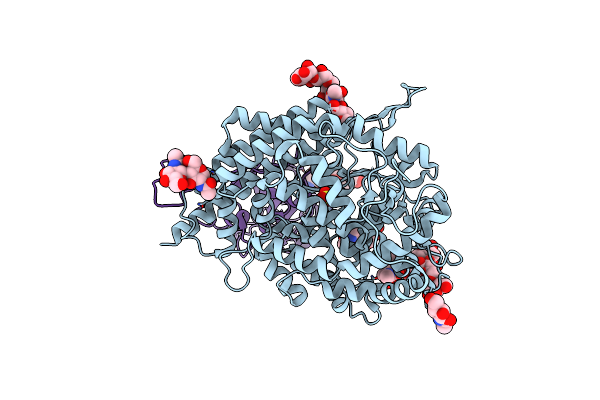 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2023-12-27 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG, ZN, SO4 |
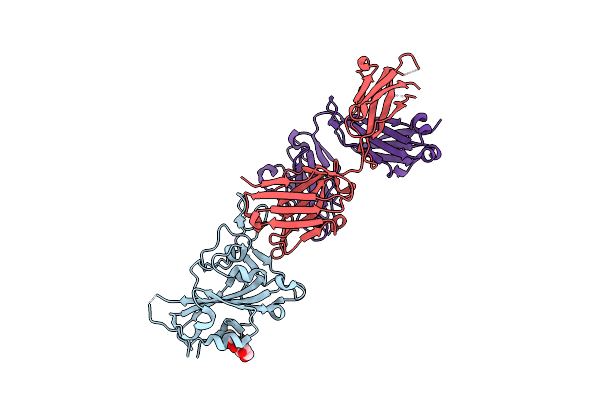 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
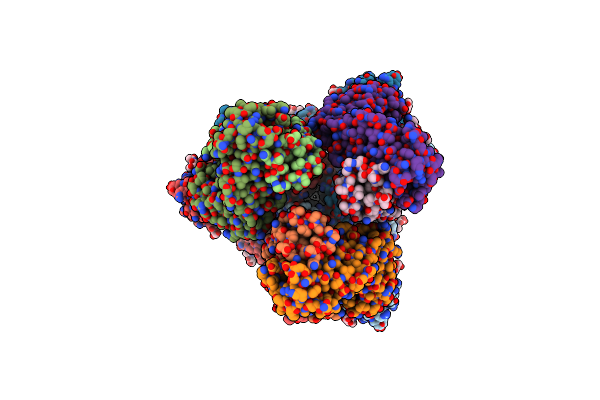 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structure Of Sars-Cov-2 Spike Rbd In Complex With Neutralizing Antibody Niv-11
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of Formate Dehydrogenase From Methylorubrum Extorquens Am1
Organism: Methylorubrum extorquens am1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-10-11 Classification: OXIDOREDUCTASE Ligands: MGD, W, FES, SF4, GOL, PEG, FMN |
 |
Neutron Structure Of [Nife]-Hydrogenase From D. Vulgaris Miyazaki F In Its Oxidized State
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.04 Å, 2.20 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: NFU, MPD, OH, MG, CL, SF4, F3S |
 |
Structure Of Sars-Cov-2 Spike Rbd In Complex With Neutralizing Antibody Niv-8
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-03-29 Classification: LYASE Ligands: CA, K, FWK, B12 |
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Release Date: 2023-03-29 Classification: LYASE Ligands: PGO, CA, K, NH4, FWK, B12 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-03-29 Classification: LYASE Ligands: GOL, NH4, FWK, B12 |
 |
Ethanolamine Ammonia-Lyase Complexed With Adomecbl In The Presence Of Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-03-29 Classification: LYASE Ligands: FWK, GOL, NH4, B12 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc, D122G/H130Y/T267C Mutant, Hydroxylamine-Treated
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2023-03-29 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc Precursor, H130Y/N266A/T267A Mutant
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: GOL, SO4, NA |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc Precursor, D122G/H130Y/T267C Mutant
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: GOL, NA, SO4 |
 |
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-01-18 Classification: STRUCTURAL PROTEIN |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2022-06-29 Classification: OXYGEN BINDING Ligands: HEM, O |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2022-06-29 Classification: OXYGEN BINDING Ligands: HEM |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2022-06-29 Classification: OXYGEN BINDING Ligands: HEM |

