Search Count: 26
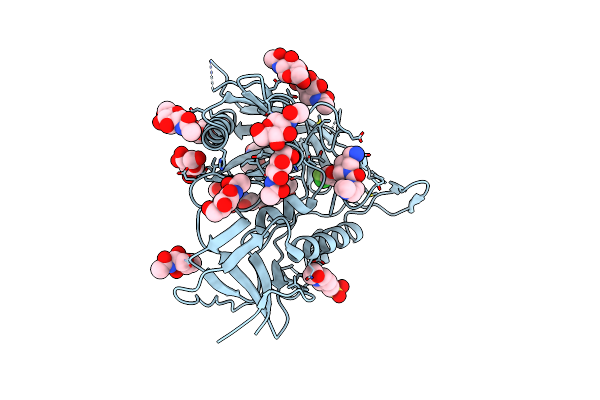 |
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Zxc-I-090
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, YZI, EPE |
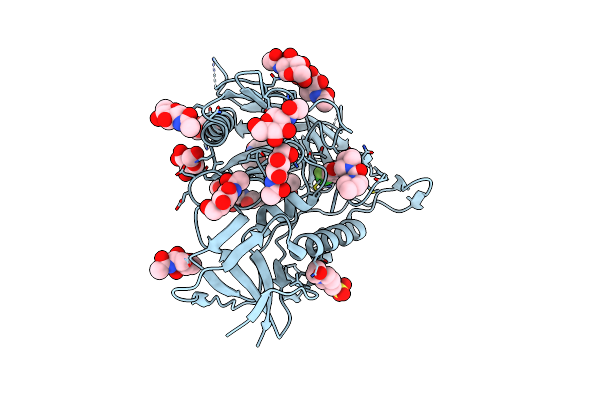 |
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Tfh-I-070-A6
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, Z3F, EPE, CL |
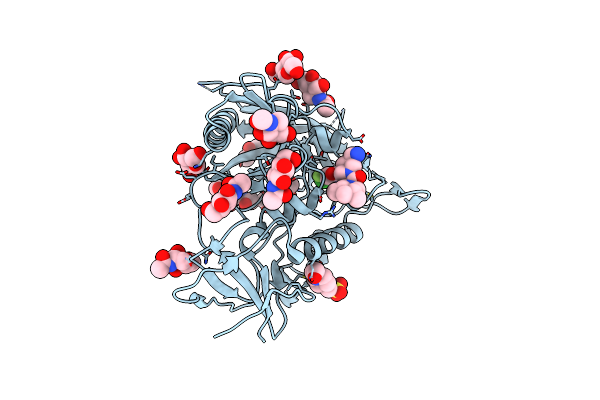 |
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Zxc-I-092
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, Z1I, EPE, CL |
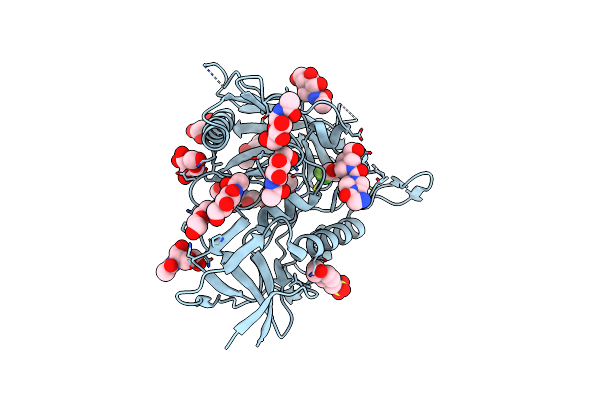 |
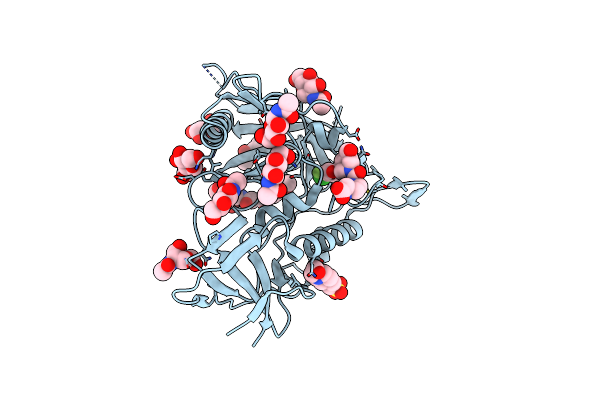
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Dl-I-101
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, EPE, Z1Z, PEG |
 |
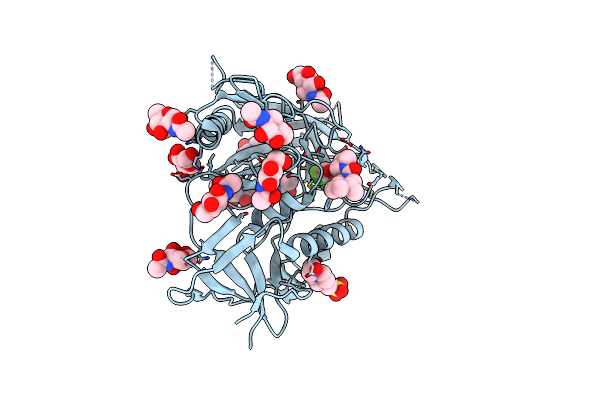
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Dl-I-102
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, EPE, Z2O |
 |
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Tfh-I-116-D1
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, ZNF, EPE, CL |
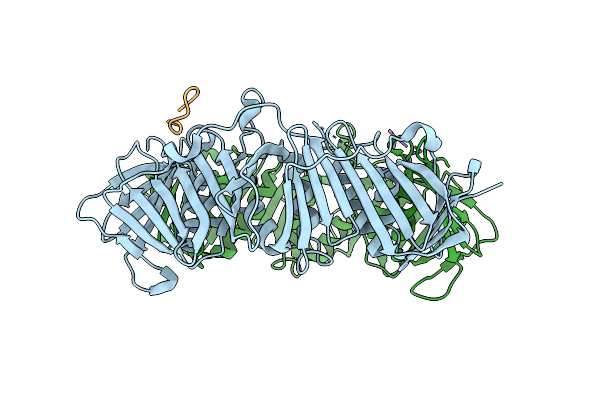 |
Structure Of Dna Polymerase Iii Subunit Beta From Rickettsia Conorii In Complex With A Natural Product
Organism: Rickettsia conorii (strain atcc vr-613 / malish 7), Streptomyces caelicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-06-12 Classification: TRANSFERASE |
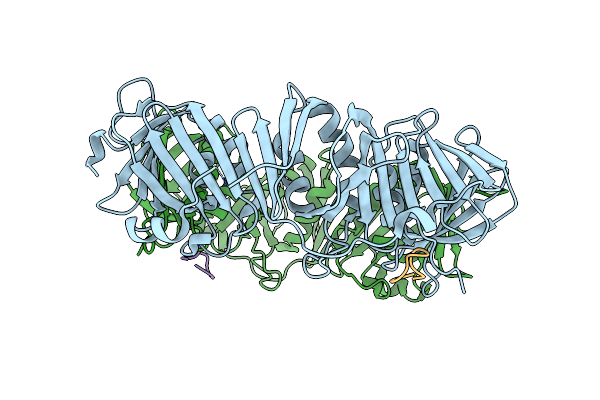 |
Structure Of Dna Polymerase Iii Subunit Beta From Borrelia Burgdorferi In Complex With A Natural Product
Organism: Borrelia burgdorferi (strain atcc 35210 / b31 / cip 102532 / dsm 4680), Streptomyces
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN |
 |
Structure Of Dna Polymerase Iii Subunit Beta From Rickettsia Typhi In Complex With A Natural Product
Organism: Rickettsia typhi str. wilmington, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN/INHIBITOR Ligands: EDO, PEG |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Naegleria Fowleri With Bound Nad
Organism: Naegleria fowleri
Method: X-RAY DIFFRACTION Release Date: 2019-01-23 Classification: OXIDOREDUCTASE Ligands: EDO, NAD |
 |
Crystal Structure Of Utp-Glucose-1-Phosphate Uridylyltransferase From Burkholderia Ambifaria
Organism: Burkholderia ambifaria (strain mc40-6)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: CIT |
 |
Crystal Structure Of Utp-Glucose-1-Phosphate Uridylyltransferase From Burkholderia Ambifaria In Complex With Utp
Organism: Burkholderia ambifaria (strain mc40-6)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: UTP |
 |
Crystal Structure Of 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase (Ispd) From Burkholderia Thailandensis Complexed With Ctp
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-29 Classification: TRANSFERASE Ligands: CTP, MG, ACT, CAD, GOL |
 |
Structure Of Complex Between The 3Rd Lim Domain Of Tes And The Evh1 Domain Of Mena
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2007-10-16 Classification: METAL-BINDING Ligands: ZN |
 |
Organism: Talaromyces emersonii
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2004-11-09 Classification: HYDROLASE Ligands: NAG |
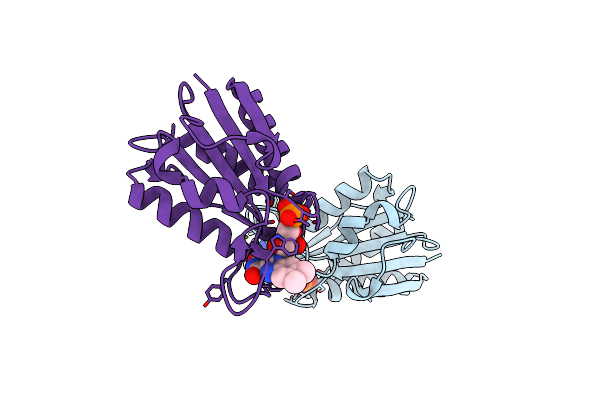 |
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2000-08-23 Classification: ELECTRON TRANSPORT Ligands: FMN |
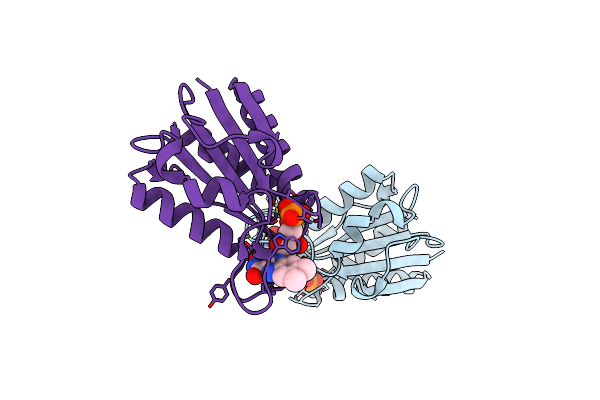 |
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-23 Classification: ELECTRON TRANSPORT Ligands: FMN |
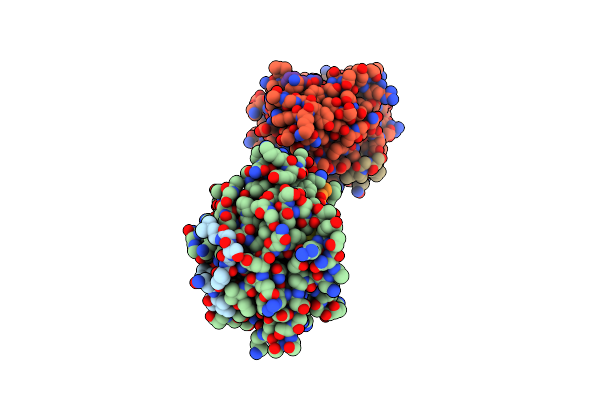 |
Crystal Structure Analysis Of Delta-Chymotrypsin Bound To A Peptidyl Chloromethyl Ketone Inhibitor
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2000-05-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CL |
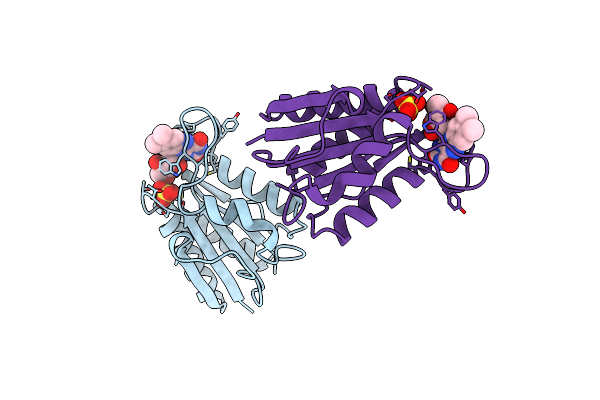 |
X-Ray Crystal Structure Of The Desulfovibrio Vulgaris (Hildenborough) Apoflavodoxin-Riboflavin Complex
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 1999-02-09 Classification: ELECTRON TRANSPORT Ligands: SO4, RBF |
 |
Organism: Desulfovibrio vulgaris subsp. vulgaris str. hildenborough
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-12-02 Classification: ELECTRON TRANSPORT Ligands: FMN |

