Search Count: 16
 |
Identification Of An Intermediate Activation State Of Pak5 Reveals A Novel Mechanism Of Kinase Inhibition.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-10-25 Classification: TRANSFERASE |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-11-03 Classification: PROTEIN BINDING,HYDROLASE Ligands: GOL, 89N |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2021-11-03 Classification: PROTEIN BINDING,HYDROLASE Ligands: 89N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-10-13 Classification: OXYGEN TRANSPORT Ligands: CMO, HEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-02-25 Classification: TRANSFERASE Ligands: AWR, PO4 |
 |
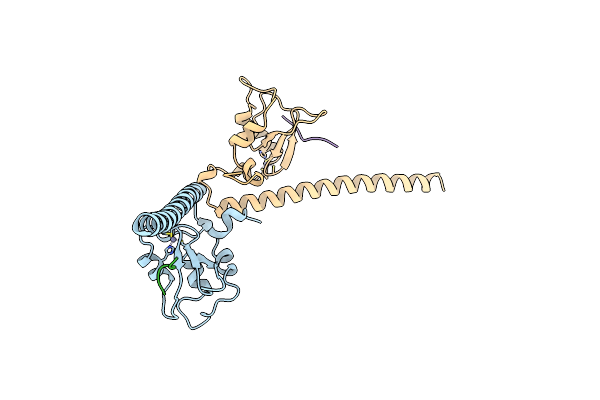
Crystal Structure Of Human Survivin Bound To Histone H3 Phosphorylated On Threonine-3.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2012-03-07 Classification: CELL CYCLE Ligands: ZN, EDO, PG4, 1PE, ACT, PEG |
 |
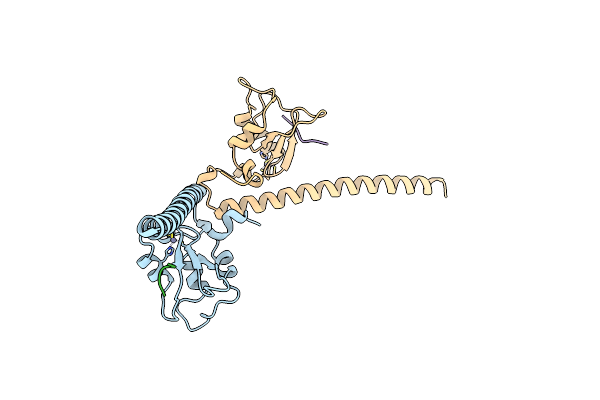
Crystal Structure Of Human Survivin Bound To Histone H3 Phosphorylated On Threonine-3 (C2 Space Group).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-03-07 Classification: CELL CYCLE Ligands: ZN |
 |
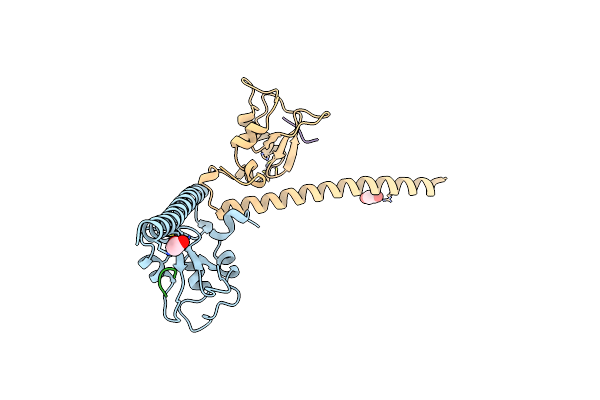
Crystal Structure Of Human Survivin K62A Mutant Bound To N-Terminal Histone H3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2012-03-07 Classification: CELL CYCLE Ligands: ZN |
 |
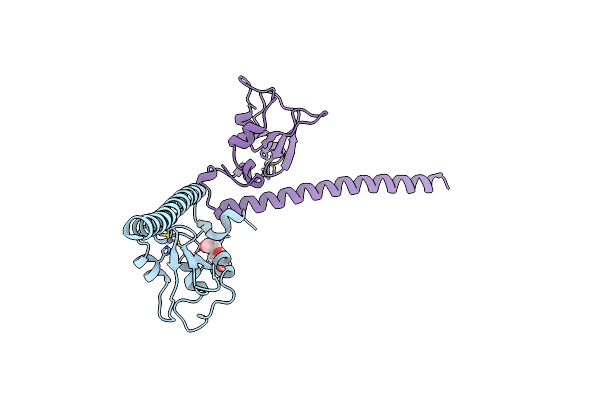
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2012-03-07 Classification: CELL CYCLE Ligands: ZN, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-03-07 Classification: CELL CYCLE Ligands: ZN, EDO, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-03-07 Classification: CELL CYCLE Ligands: ZN, EDO, PEG, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-03-07 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-09-08 Classification: TRANSFERASE Ligands: PO4, 5ID, IOD, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-09-16 Classification: TRANSFERASE Ligands: IOD, 5ID, NI, DMS, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-07-15 Classification: TRANSFERASE Ligands: MG, AMP, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-09 Classification: CELL ADHESION, METAL BINDING PROTEIN Ligands: CA |

