Search Count: 60
 |
X-Ray Crystal Structure Of Asp/Ala Exchanger Aspt At Outward-Facing Conformation
Organism: Tetragenococcus halophilus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: TRANSPORT PROTEIN Ligands: ASP |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: TRANSPORT PROTEIN |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: MG, NOJ |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: MG, NOJ |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: TRANSPORT PROTEIN Ligands: ASP |
 |
Organism: Tetragenococcus halophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Gh42 Beta-Galactosidase Bibga42A From Bifidobacterium Longum Subspecies Infantis In Complex With Glycerol
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: GOL, PEG |
 |
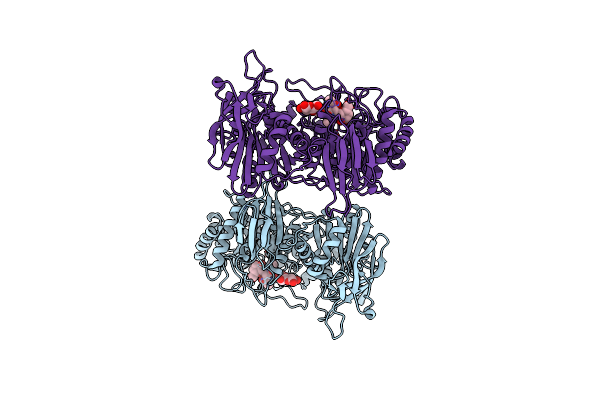
Crystal Structure Of Gh42 Beta-Galactosidase Bibga42A From Bifidobacterium Longum Subspecies Infantis E160A/E318A Mutant In Complex With Galactose
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: GLA |
 |
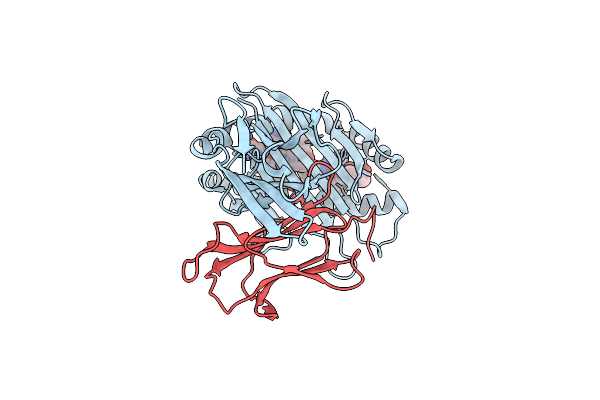
Crystal Structure Of Gh42 Beta-Galactosidase Bibga42A From Bifidobacterium Longum Subspecies Infantis E318S Mutant In Complex With Lacto-N-Tetraose
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-07 Classification: HYDROLASE |
 |
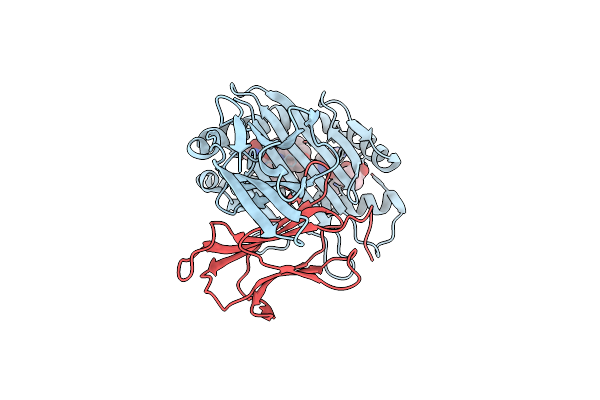
Complex Structure Of Hla-A*2402 With The Peptide From Hcov(Cov-2) Spike Protein
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-01-26 Classification: IMMUNE SYSTEM |
 |
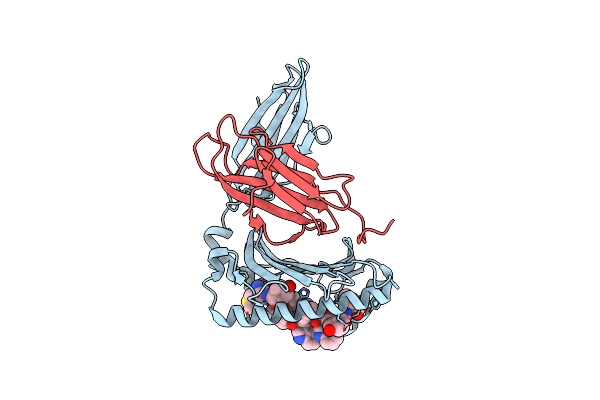
Complex Structure Of Hla-A*2402 With The Peptide From Hcov(Cov-229E) Spike Protein
Organism: Homo sapiens, Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-01-26 Classification: IMMUNE SYSTEM |
 |
Complex Structure Of Hla-A*2402 With The Peptide From Hcov(Cov-Hku1) Spike Protein
Organism: Homo sapiens, Human coronavirus hku1
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-01-26 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Galacto-N-Biose/Lacto-N-Biose I Phosphorylase C236Y Mutant
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-10-09 Classification: TRANSFERASE Ligands: NDG |
 |
Crystal Structure Of Alpha-1,3/4-Fucosidase From Bifidobacterium Longum Subsp. Infantis Complexed With Deoxyfuconojirimycin
Organism: Bifidobacterium longum subsp. infantis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-04-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DFU, EDO, NA |
 |
Crystal Structure Of Alpha-1,3/4-Fucosidase From Bifidobacterium Longum Subsp. Infantis D172A/E217A Mutant Complexed With Lacto-N-Fucopentaose Ii
Organism: Bifidobacterium longum subsp. infantis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-04-04 Classification: HYDROLASE |
 |
Crystal Structure Of The Carboxy-Terminal Ribonuclease Domain Of Colicin E5 R33Q Mutant
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-11-02 Classification: HYDROLASE Ligands: 3PD, UM3 |
 |
Crystal Structure Of The C-Terminal Domain Of Sequence-Specific Ribonuclease
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-09-28 Classification: HYDROLASE Ligands: CD |
 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Complexed With Sulfate And Isofagomine
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-09-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BGC, SO4, IFM, EPE |
 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Complexed With Sulfate And 1-Deoxynojirimycin
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2011-09-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BGC, SO4, NOJ, EPE |

