Search Count: 22
 |
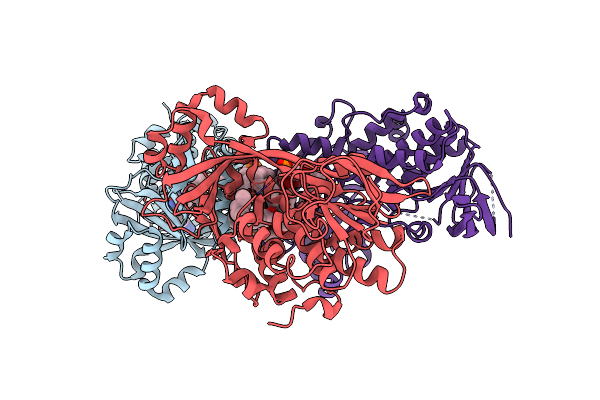
Cryo-Em Structure Of Burkholderia Cenocepacia Orotate Phosphoribosyltransferase
Organism: Burkholderia cenocepacia j2315
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: TRANSFERASE Ligands: ACT |
 |
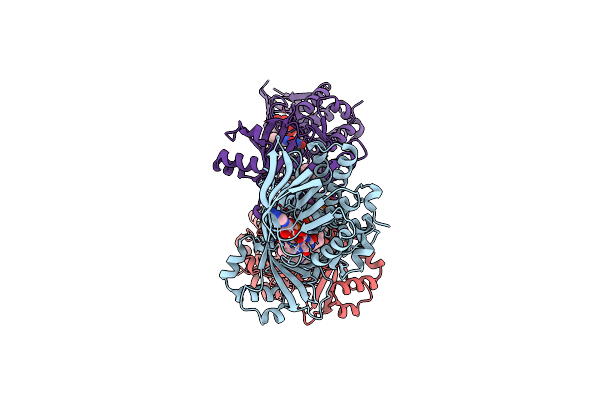
Organism: Neobacillus niacini
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: FAD, DR9 |
 |
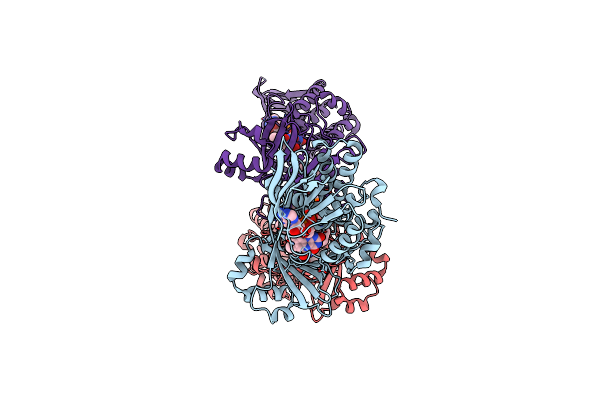
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, DR9 |
 |
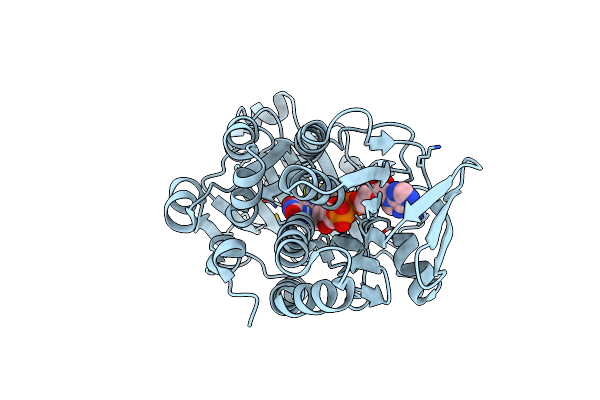
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, WTQ, DR9 |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: FAD, PYS |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2024-02-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: VJX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: VXE |
 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2019-04-03 Classification: LYASE Ligands: ACT, 3HB |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-01-23 Classification: TRANSFERASE Ligands: PO4, CL |
 |
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-12-13 Classification: HYDROLASE Ligands: PO4, K |
 |
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-12-13 Classification: HYDROLASE Ligands: AG2, K |
 |
Crystal Structure Of A Short Chain Dehydrogenase From Burkholderia Cenocepacia J2315 In Complex With Nadp.
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Release Date: 2017-01-18 Classification: OXIDOREDUCTASE Ligands: NAP, BEZ, PO4 |
 |
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-06-29 Classification: TRANSFERASE |
 |
Crystal Structure Of 6-Hydroxynicotinic Acid 3-Monooxygenase From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida (strain kt2440)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-06-08 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Structure Of A Putative Phosphomethylpyrimidine Kinase From Acinetobacter Baumannii In Non-Covalent Complex With Pyridoxal Phosphate
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Brucella ovis intabari-2002-82-58
Method: X-RAY DIFFRACTION Release Date: 2016-02-10 Classification: OXIDOREDUCTASE Ligands: IMD, EDO, NAD |
 |
Organism: Brucella ovis (strain atcc 25840 / 63/290 / nctc 10512)
Method: X-RAY DIFFRACTION Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: ACT, EDO |
 |
Structure Of A Putative Phosphomethylpyrimidine Kinase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii is-123
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-04-01 Classification: TRANSFERASE Ligands: EDO |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-05-16 Classification: OXIDOREDUCTASE Ligands: FAD |

