Search Count: 17
 |
Organism: Lachancea thermotolerans (strain atcc 56472 / cbs 6340 / nrrl y-8284)
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2022-09-07 Classification: STRUCTURAL PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-09-07 Classification: STRUCTURAL PROTEIN Ligands: JEF |
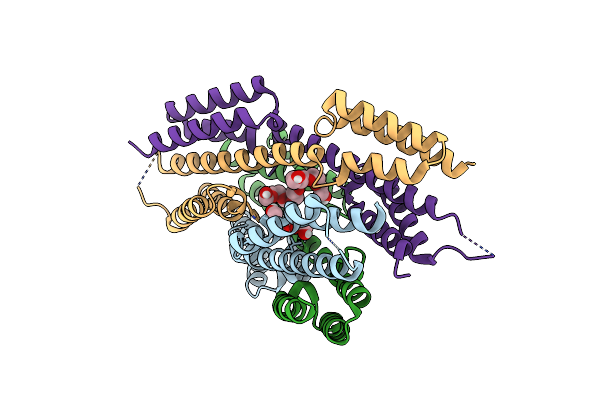 |
Crystal Structure Of The Dimeric Mitofilin Domain Of Mic60 In Complex With The Chch Domain Of Mic19
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-09-07 Classification: STRUCTURAL PROTEIN Ligands: PG4 |
 |
Structure Of S-Mgm1 Decorating The Outer Surface Of Tubulated Lipid Membranes
Organism: Chaetomium thermophilum
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
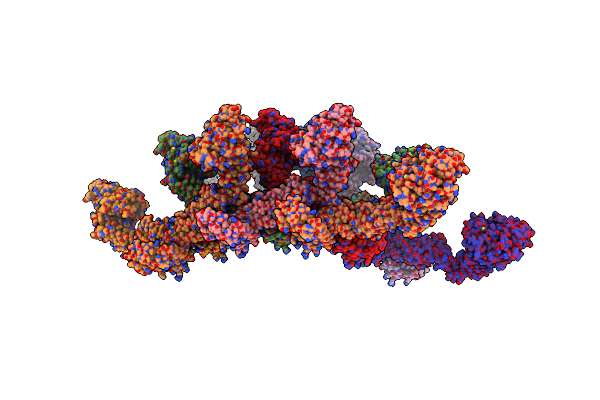 |
Structure Of S-Mgm1 Decorating The Outer Surface Of Tubulated Lipid Membranes In The Gtpgammas Bound State
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
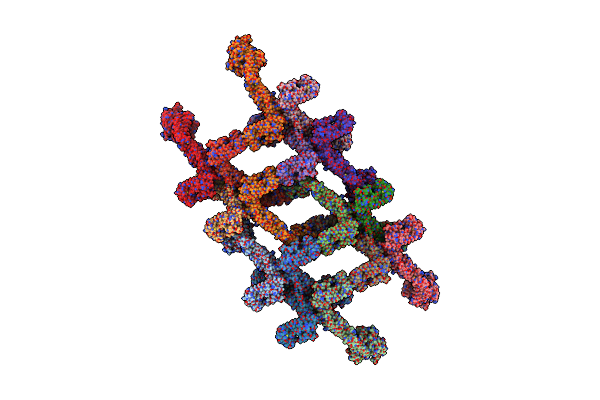 |
Structure Of S-Mgm1 Decorating The Inner Surface Of Tubulated Lipid Membranes
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
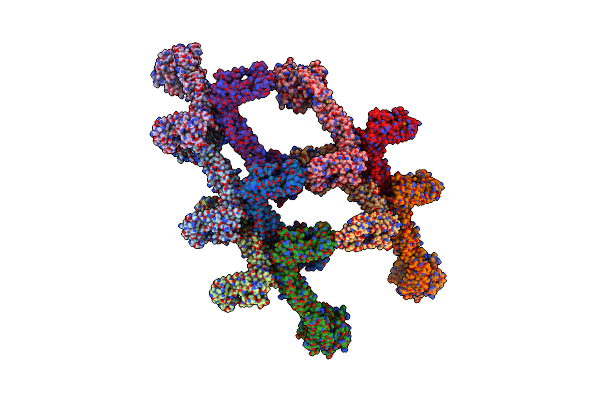 |
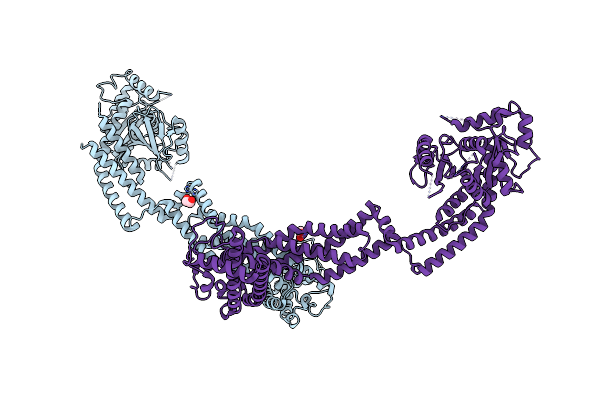
Structure Of S-Mgm1 Decorating The Inner Surface Of Tubulated Lipid Membranes In The Gtpgammas Bound State
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2019-07-03 Classification: MOTOR PROTEIN Ligands: EDO |
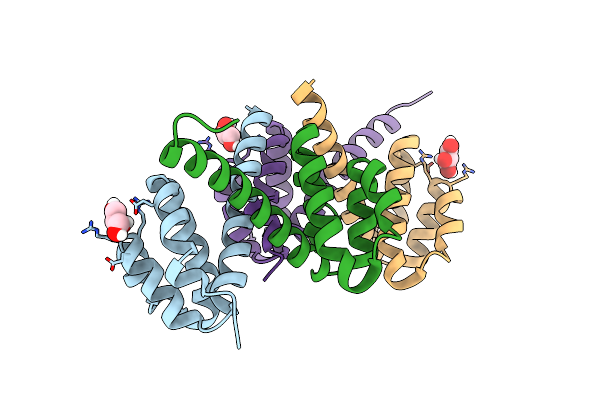 |
Crystal Structures Of Nlrp14 Pyrin Domain Reveal A Conformational Switch Mechanism, Regulating Its Molecular Interactions
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-07-16 Classification: SIGNALING PROTEIN Ligands: GOL |
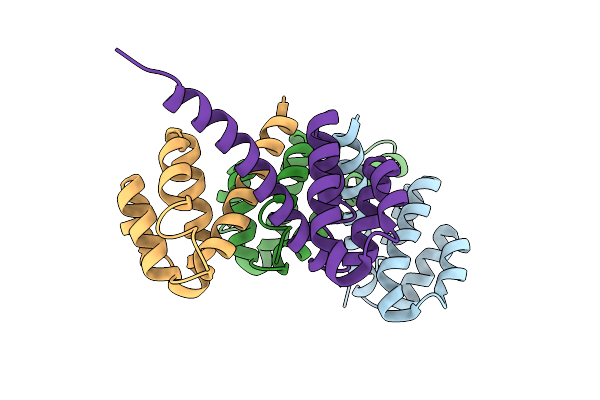 |
Crystal Structures Of Nlrp14 Pyrin Domain Reveal A Conformational Switch Mechanism, Regulating Its Molecular Interactions
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-07-16 Classification: SIGNALING PROTEIN |
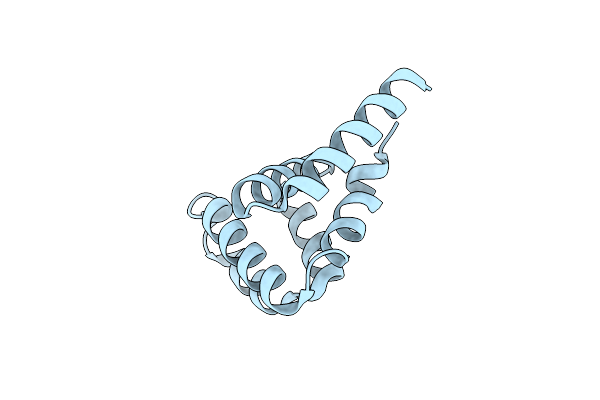 |
Crystal Structures Of Nlrp14 Pyrin Domain Reveal A Conformational Switch Mechanism, Regulating Its Molecular Interactions
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-07-16 Classification: SIGNALING PROTEIN |
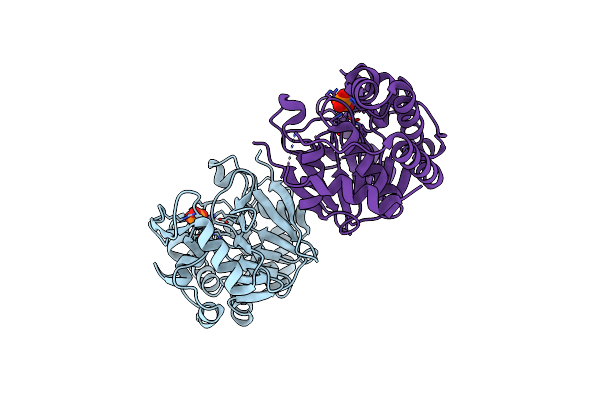 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-03-26 Classification: HYDROLASE Ligands: MN, PO4, CL |
 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-03-26 Classification: HYDROLASE/NUCLEAR PROTEIN Ligands: MN, CL, PO4, GOL |
 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-26 Classification: HYDROLASE Ligands: MN, GOL, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: FLC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: GNP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2012-09-05 Classification: SIGNALING PROTEIN, PROTEIN BINDING Ligands: CL, SO4 |

