Search Count: 27
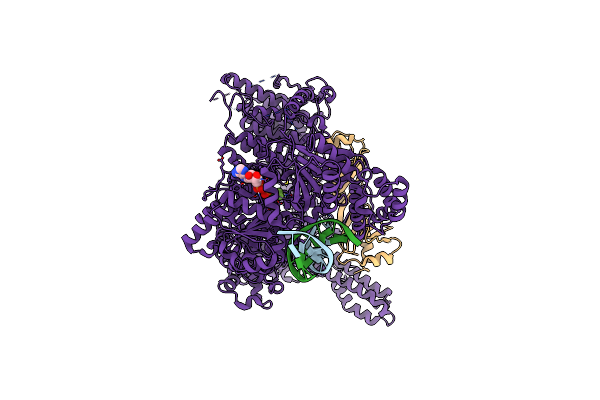 |
Organism: Chaetomium thermophilum, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: TRANSCRIPTION Ligands: MG, ADP, BEF |
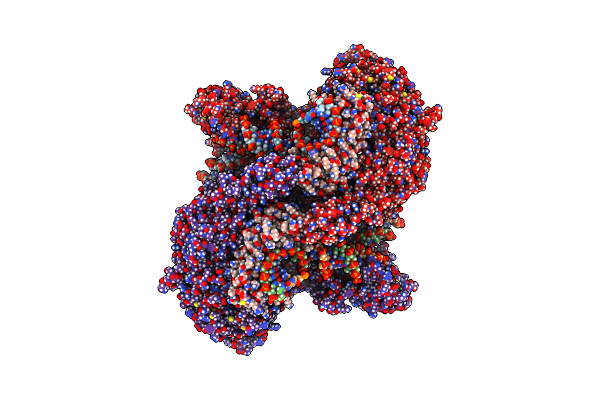 |
Organism: Chaetomium thermophilum, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: TRANSCRIPTION |
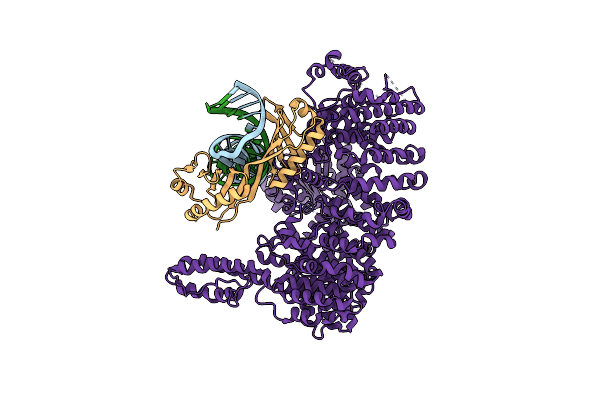 |
Organism: Chaetomium thermophilum, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSCRIPTION |
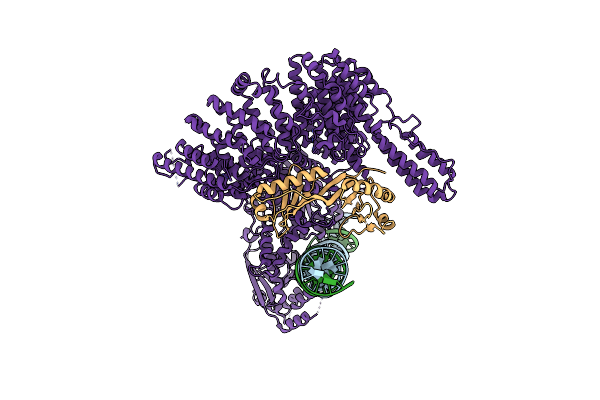 |
Organism: Chaetomium thermophilum, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-10-28 Classification: RIBOSOME Ligands: MG, ZN, ADP, ATP, SF4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-10-14 Classification: RIBOSOME Ligands: ZN, SF4, ADP, MG, ATP, MET, GTP |
 |
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-10-14 Classification: RIBOSOME Ligands: MG, ZN, SF4, ADP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2020-10-07 Classification: RIBOSOME Ligands: ZN, ADP, MG, ATP, SF4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-10-07 Classification: RIBOSOME Ligands: ZN, SF4, ADP, MG, ATP |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-02-27 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-02-27 Classification: TRANSPORT PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-08-01 Classification: STRUCTURAL PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-08-01 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2017-06-14 Classification: PROTEIN BINDING |
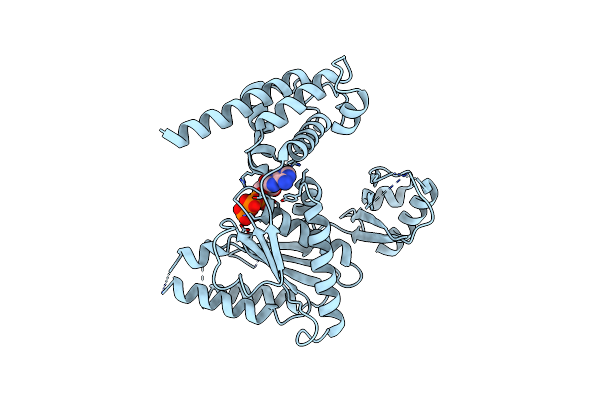 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2017-06-14 Classification: PROTEIN BINDING Ligands: ATP |
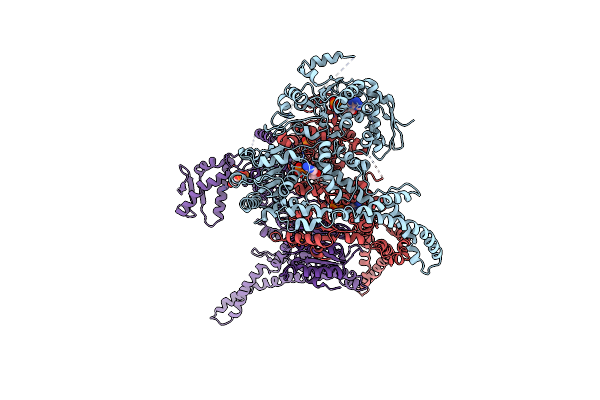 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2016-12-07 Classification: CHAPERONE Ligands: ADP |
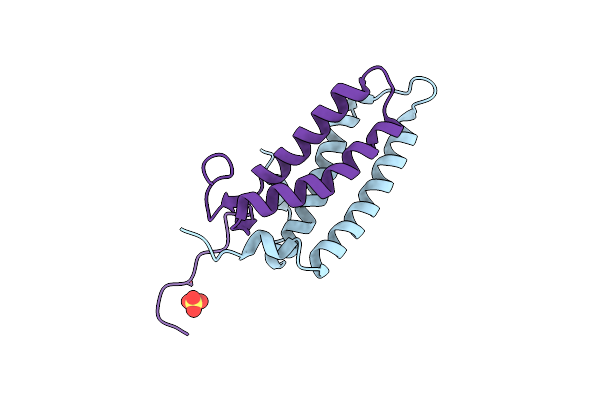 |
Crystal Structure Of The Kinetochore Mis12 Complex Head2 Subdomain Containing Dsn1 And Nsl1 Fragments
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-11-16 Classification: CELL CYCLE Ligands: SO4 |
 |
Crystal Structure Of The Human Kinetochore Mis12-Cenp-C Delta-Head2 Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2016-11-16 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-11-16 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-06-22 Classification: TRANSCRIPTION Ligands: CL |

