Search Count: 20
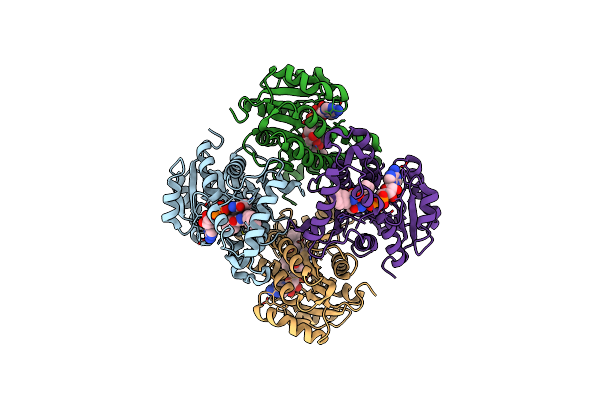 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE Ligands: NAD, 3KX |
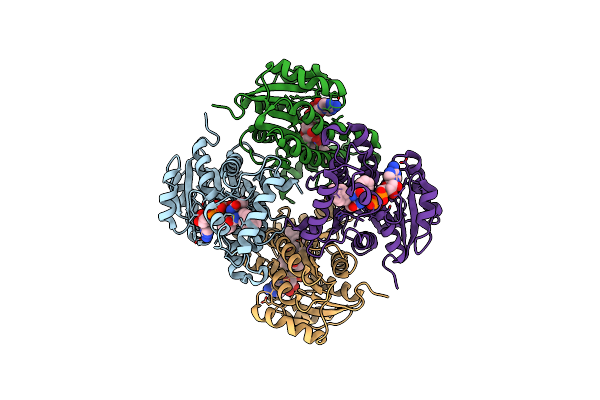 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE Ligands: NAD, 3KY |
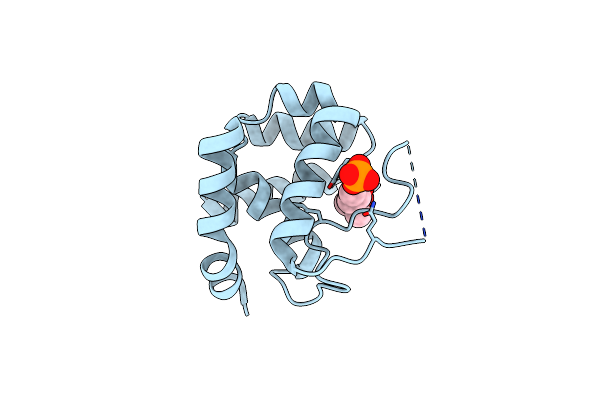 |
Crystal Structure Of The Endo-Beta-N-Acetylglucosaminidase From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-11-12 Classification: HYDROLASE Ligands: PO4, MPD |
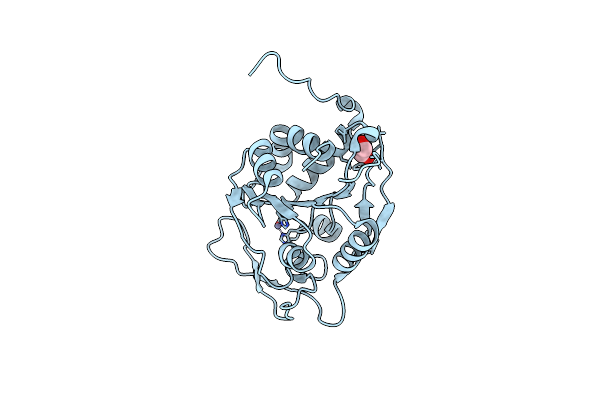 |
Biochemical And Structural Characterization Of The Mpaa Amidase As Part Of A Conserved Scavenging Pathway For Peptidoglycan Derived Peptides In Gamma-Proteobacteria
Organism: Vibrio campbellii atcc baa-1116
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2012-09-26 Classification: HYDROLASE Ligands: ZN, EDO |
 |
The Structure Of The Escherichia Coli Murein Tripeptide Binding Protein Mppa
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2011-07-06 Classification: PEPTIDE BINDING PROTEIN/PEPTIDE Ligands: MHI, ZN |
 |
Crystal Structure Of The Three-Pasta-Domain Of A Ser/Thr Kinase From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-11-03 Classification: TRANSFERASE Ligands: ZN |
 |
Crystal Structure Of A Murein Peptide Ligase Mpl (Psyc_0032) From Psychrobacter Arcticus 273-4 At 1.65 A Resolution
Organism: Psychrobacter arcticus 273-4
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-06-16 Classification: LIGASE |
 |
Crystal Structure Of A Ld-Carboxypeptidase A (Saro_1426) From Novosphingobium Aromaticivorans Dsm At 1.89 A Resolution
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2009-02-10 Classification: HYDROLASE Ligands: SO4, GOL, UNL |
 |
Crystal Structure Of Murd Ligase In Complex With D-Glu Containing Sulfonamide Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2007-05-15 Classification: LIGASE Ligands: LK2, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2007-05-15 Classification: LIGASE Ligands: UMA, ADP, SO4 |
 |
Crystal Structure Of Murd Ligase In Complex With L-Glu Containing Sulfonamide Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2007-05-15 Classification: LIGASE Ligands: LK1, SO4 |
 |
Crystal Structure Of The Y10F Mutant Of The Gluathione S-Transferase From Schistosoma Haematobium
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-07-04 Classification: TRANSFERASE Ligands: GSH |
 |
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2006-06-26 Classification: TRANSFERASE Ligands: GSH, PG4 |
 |
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-06-21 Classification: TRANSFERASE Ligands: GTX, PG4 |
 |
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-06-21 Classification: TRANSFERASE Ligands: SO4, BME |
 |
Structure Of Glutathione-S-Transferase Mutant, R21L, From Schistosoma Haematobium
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2006-06-21 Classification: TRANSFERASE Ligands: SO4, PG4, BME |
 |
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-06-21 Classification: TRANSFERASE Ligands: GSH, PG4, BME |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-09-14 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-07-25 Classification: TRANSFERASE Ligands: GSH |
 |
28Kda Glutathione S-Transferase From Schistosoma Haematobium (Glutathione Saturated)
Organism: Schistosoma haematobium
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2003-07-25 Classification: TRANSFERASE Ligands: GSH |

