Search Count: 15
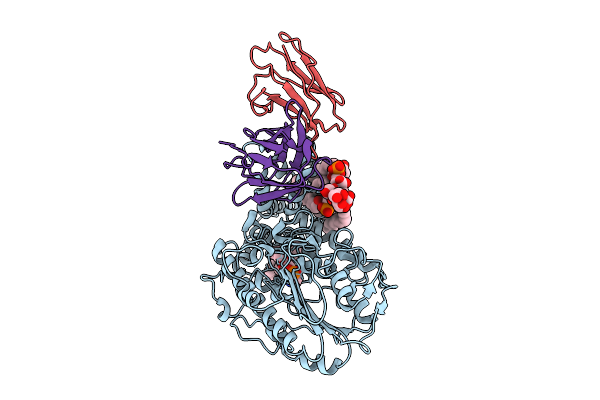 |
Single-Particle Cryo-Em Structure Of The First Variant Of Mobilized Colistin Resistance (Mcr-1) In Its Ligand-Bound State
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: KDL, PEE |
 |
Cryo-Em Structure Of Human Dynactin Complex Bound To Chlamydia Effector Dre1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MOTOR PROTEIN Ligands: ADP, ANP, ZN |
 |
Cryo-Em Structure Of Human Dynactin Complex Bound To Chlamydia Effector Dre1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MOTOR PROTEIN Ligands: ADP, ANP, ZN |
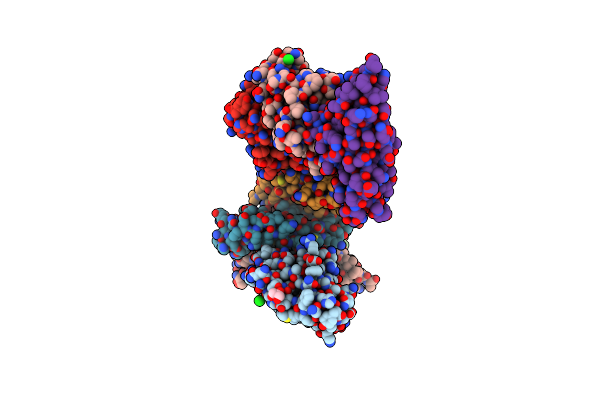 |
Organism: Homo sapiens, Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2023-03-15 Classification: CELL ADHESION Ligands: GOL, SO4, CL |
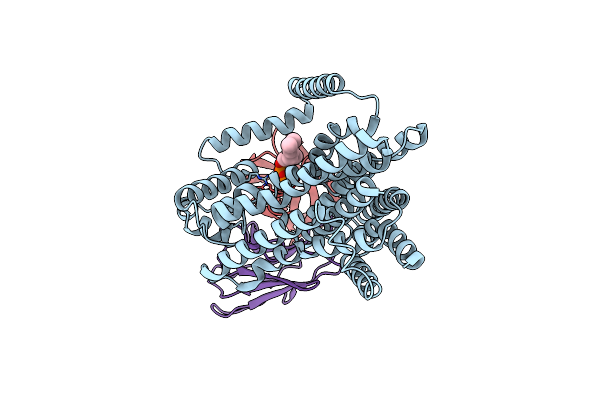 |
Single-Particle Cryo-Em Structure Of The Waal O-Antigen Ligase In Its Ligand Bound State
Organism: Cupriavidus metallidurans, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: GPP |
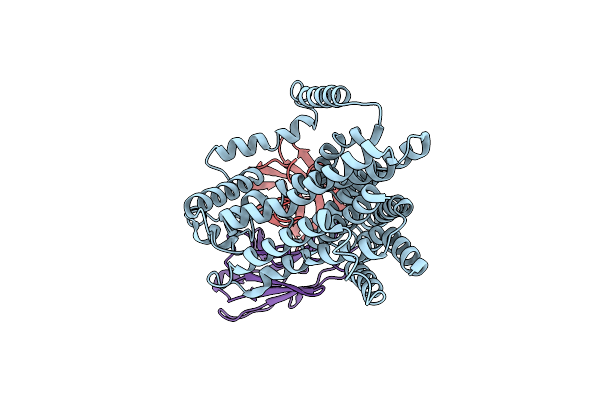 |
Single-Particle Cryo-Em Structure Of The Waal O-Antigen Ligase In Its Apo State
Organism: Cupriavidus metallidurans, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-02-13 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-02-13 Classification: SIGNALING PROTEIN Ligands: MG |
 |
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: ZN, DSL, MPG, CL, PO4, PC, EPE |
 |
Crystal Structure Of Arnt From Cupriavidus Metallidurans Bound To Undecaprenyl Phosphate
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: MPG, EPE, PO4, CL, 5TR |
 |
Organism: Ricinus communis, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-06-11 Classification: HYDROLASE/IMMUNE SYSTEM Ligands: CL, EDO |
 |
Organism: Ricinus communis, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-06-11 Classification: HYDROLASE/IMMUNE SYSTEM Ligands: ZN, ACY, CL, EDO |
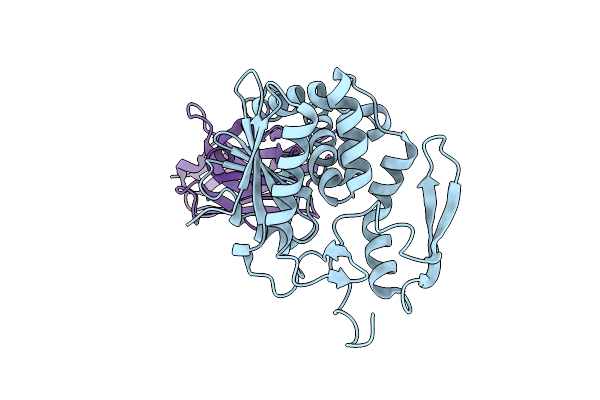 |
Organism: Ricinus communis, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-06-11 Classification: HYDROLASE/IMMUNE SYSTEM |
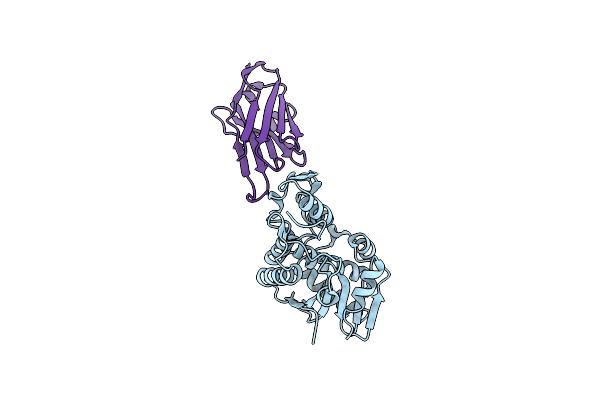 |
Organism: Ricinus communis, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-06-11 Classification: HYDROLASE/IMMUNE SYSTEM Ligands: CL |
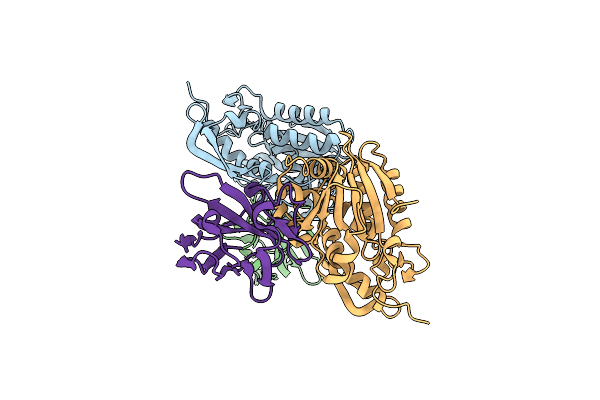 |
Organism: Ricinus communis, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-06-11 Classification: HYDROLASE/IMMUNE SYSTEM |

