Search Count: 160
 |
Organism: Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-22 Classification: DNA Ligands: MG, CL, HT1 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-06-05 Classification: TRANSFERASE Ligands: HVH, GOL, MG, PO4 |
 |
Organism: Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-01-18 Classification: DNA Ligands: O89 |
 |
Organism: Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2023-01-18 Classification: DNA |
 |
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-11-23 Classification: TRANSCRIPTION Ligands: GOL, NAD, K |
 |
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-11-09 Classification: TRANSCRIPTION |
 |
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-11-09 Classification: TRANSCRIPTION Ligands: ATP, K, NA |
 |
Organism: Agrobacterium fabrum (strain c58 / atcc 33970), Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:4.30 Å Release Date: 2022-11-09 Classification: TRANSCRIPTION |
 |
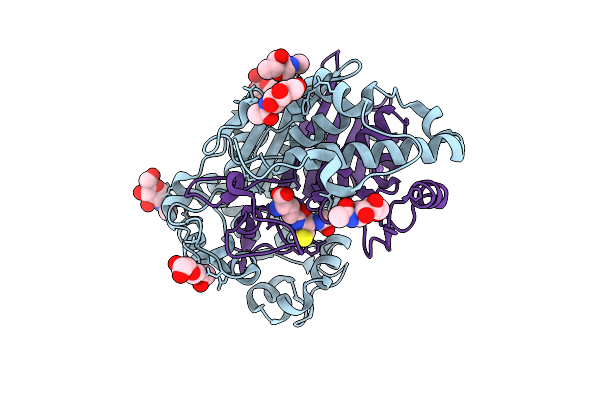
Structure Of Human Ggt1 In Complex With 2-Amino-4-(((1-((Carboxymethyl)Amino)-1-Oxobutan-2-Yl)Oxy)(Phenoxy)Phosphoryl)Butanoic Acid (Acpb) Molecule
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2020-11-25 Classification: HYDROLASE Ligands: NAG, CL, NA, V7D |
 |
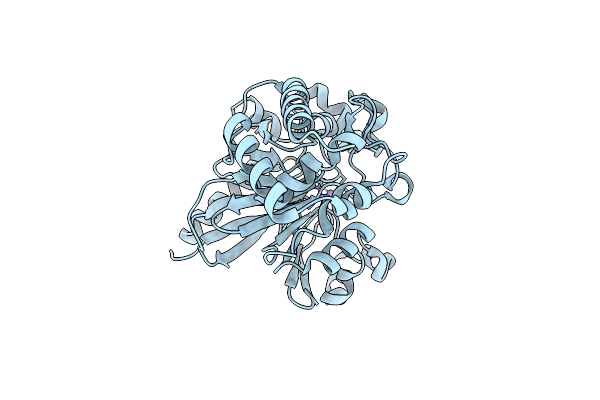
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-11-25 Classification: HYDROLASE Ligands: NAG, CL, NA, GSH |
 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ZN, MN |
 |
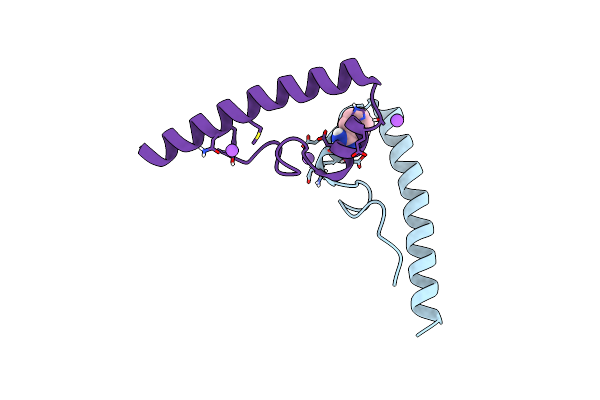
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-04-19 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAG, CL, NA, 8WY |
 |
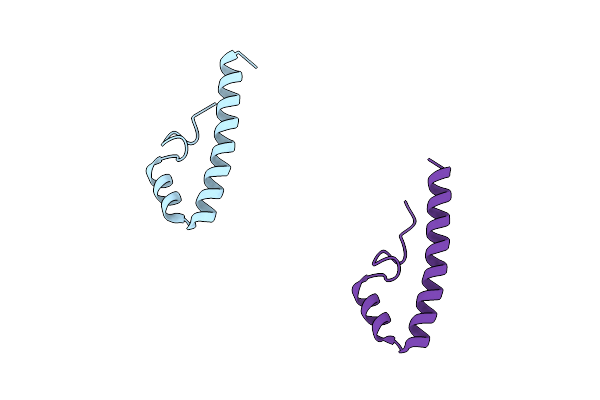
Crystal Structure Of The S. Cerevisiae Rtf1 Histone Modification Domain Mutant R126A
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2016-11-09 Classification: TRANSCRIPTION |
 |
Crystal Structure Of The S. Cerevisiae Rtf1 Histone Modification Domain Mutant R124A R126A R128A
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-10-26 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2016-10-19 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.57 Å Release Date: 2016-10-19 Classification: PROTEIN BINDING Ligands: CL |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Delta+Phs L25K/I92F At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-22 Classification: HYDROLASE Ligands: CA, THP |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Delta+Phs L25K/I92A At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-05-11 Classification: HYDROLASE Ligands: THP, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-03-09 Classification: ISOMERASE |
 |
The First Structure Of A Full-Length Mammalian Phenylalanine Hydroxylase Reveals The Architecture Of An Auto-Inhibited Tetramer
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-02-17 Classification: OXIDOREDUCTASE Ligands: FE |

