Search Count: 92
 |
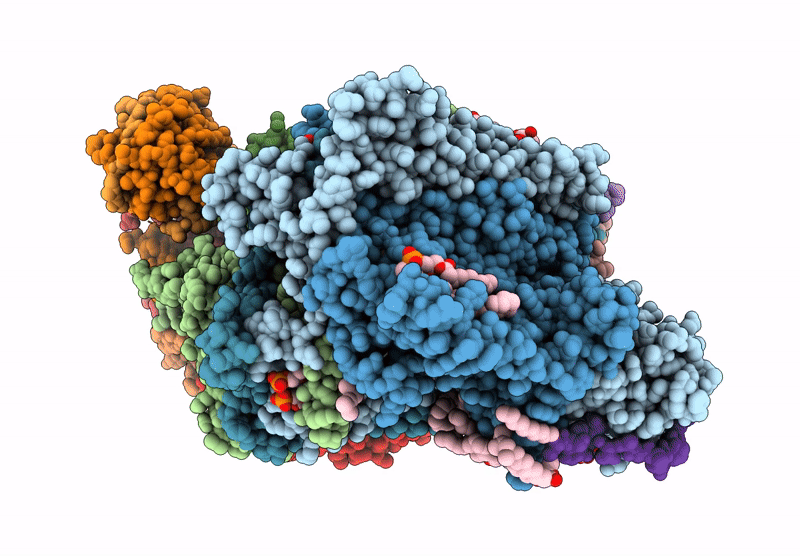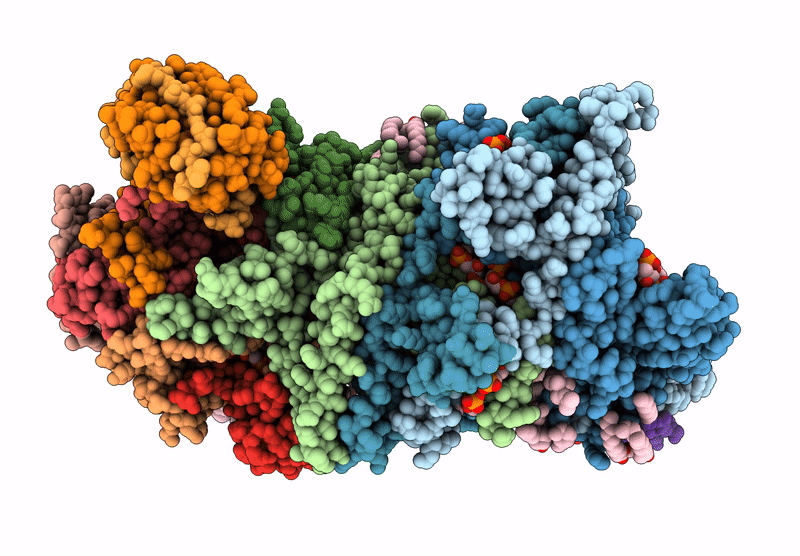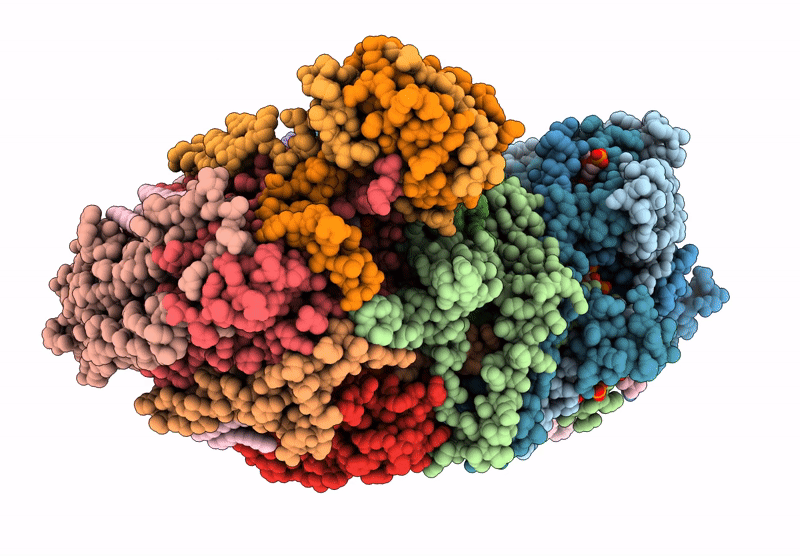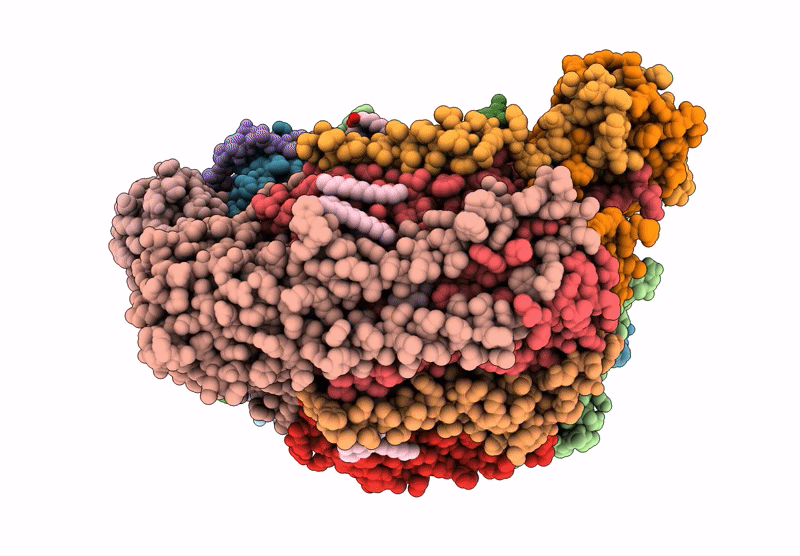
Ciii2/Civ Respiratory Supercomplex From Mycobacterium Smegmatis With Lansoprazole Sulfide
|
 |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: MEMBRANE PROTEIN Ligands: W92, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: MEMBRANE PROTEIN Ligands: Z99 |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: MEMBRANE PROTEIN Ligands: JIX, CA |
 |
Mglur3 In The Presence Of The Antagonist Ly 341495 And Positive Allosteric Modulator Vu6023326
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: MEMBRANE PROTEIN Ligands: Z99 |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: MEMBRANE PROTEIN Ligands: Z99 |
 |
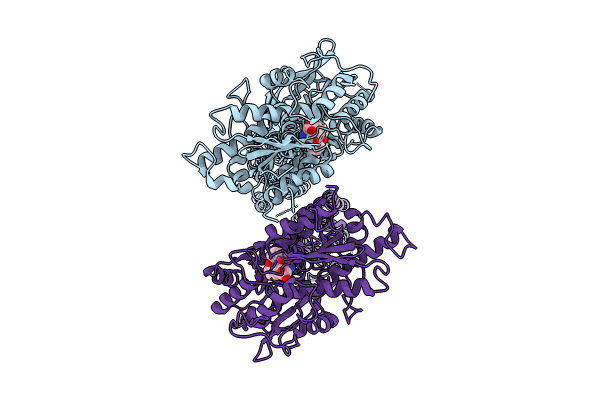
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: ELECTRON TRANSPORT Ligands: HEC, MQ9, WUO, FES, 9YF, HEM, CDL, 7PH, HEA, CU, MG, CA, 9XX, PLM |
 |
 |
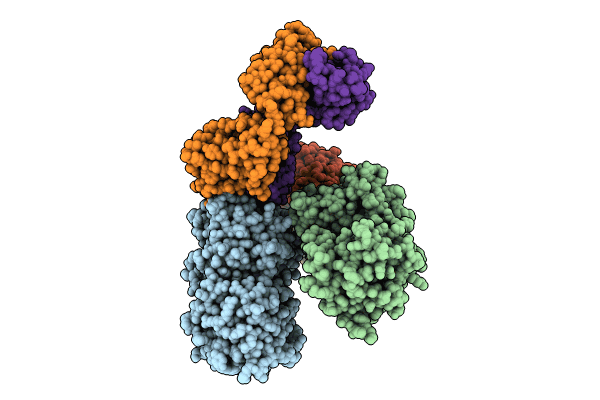
Complex Of Npr1 Ectodomain With Anp Plus An Allosteric Activating Antibody, Regn5381
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
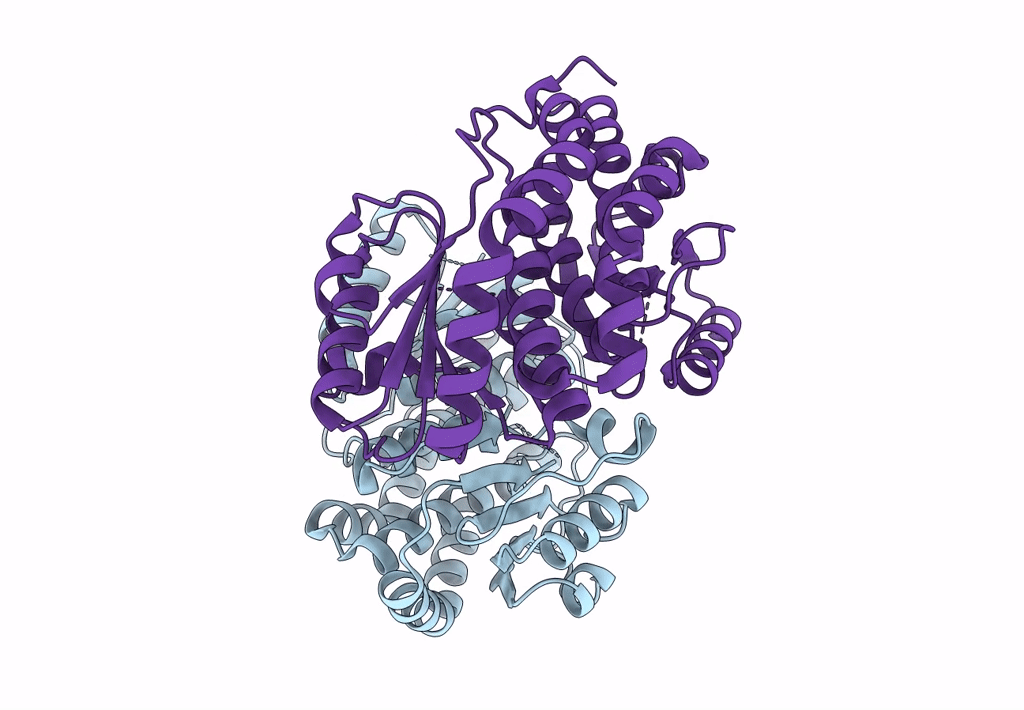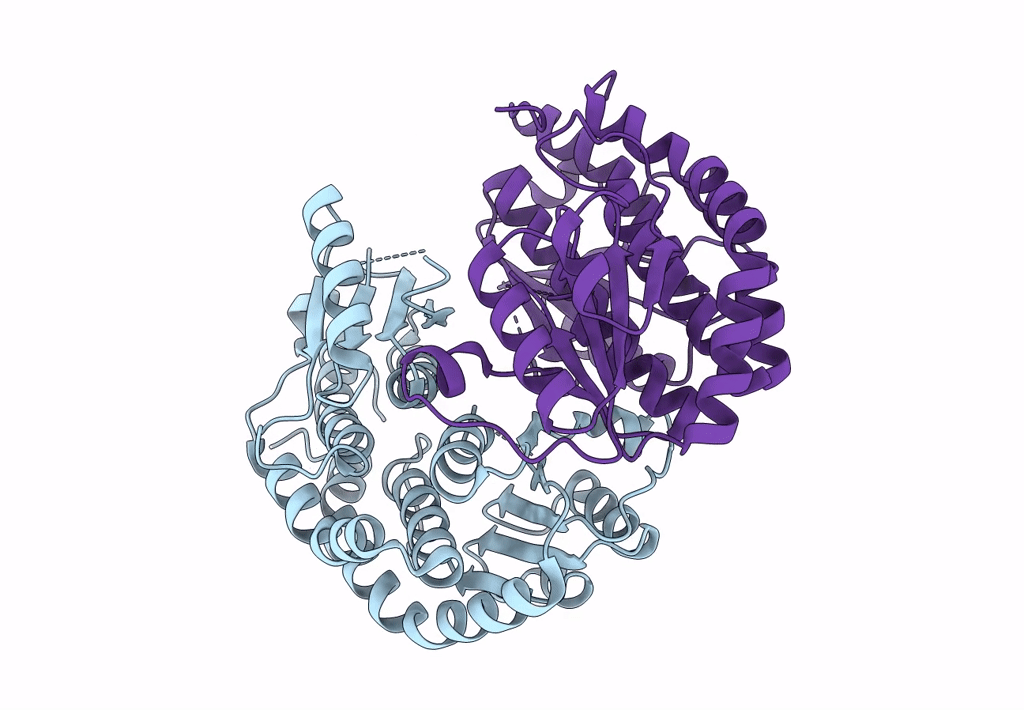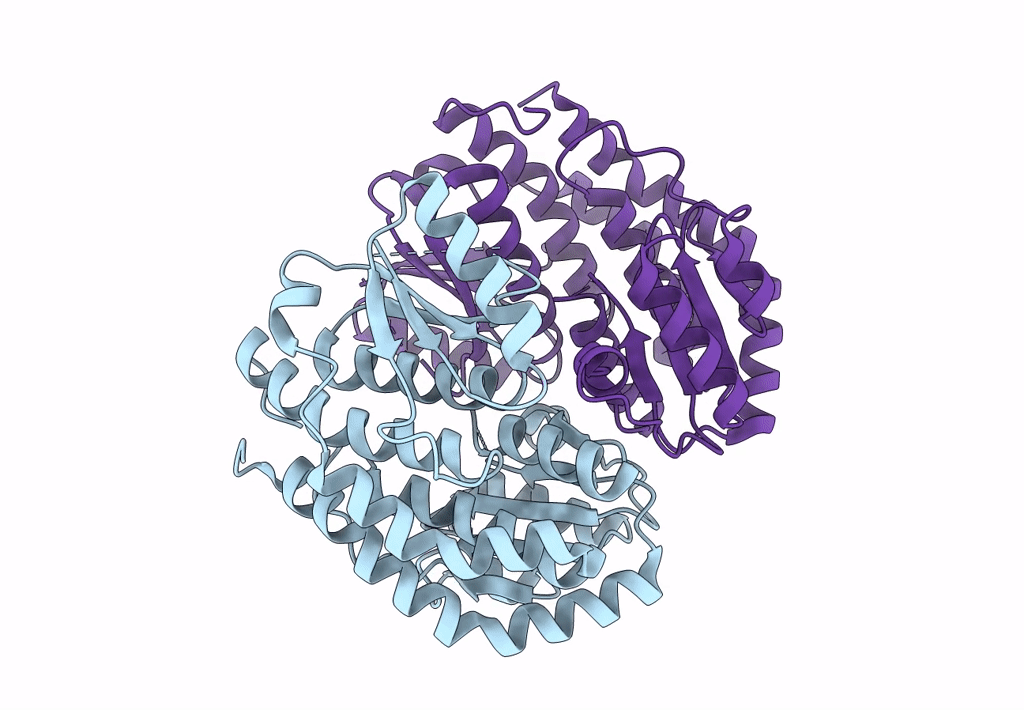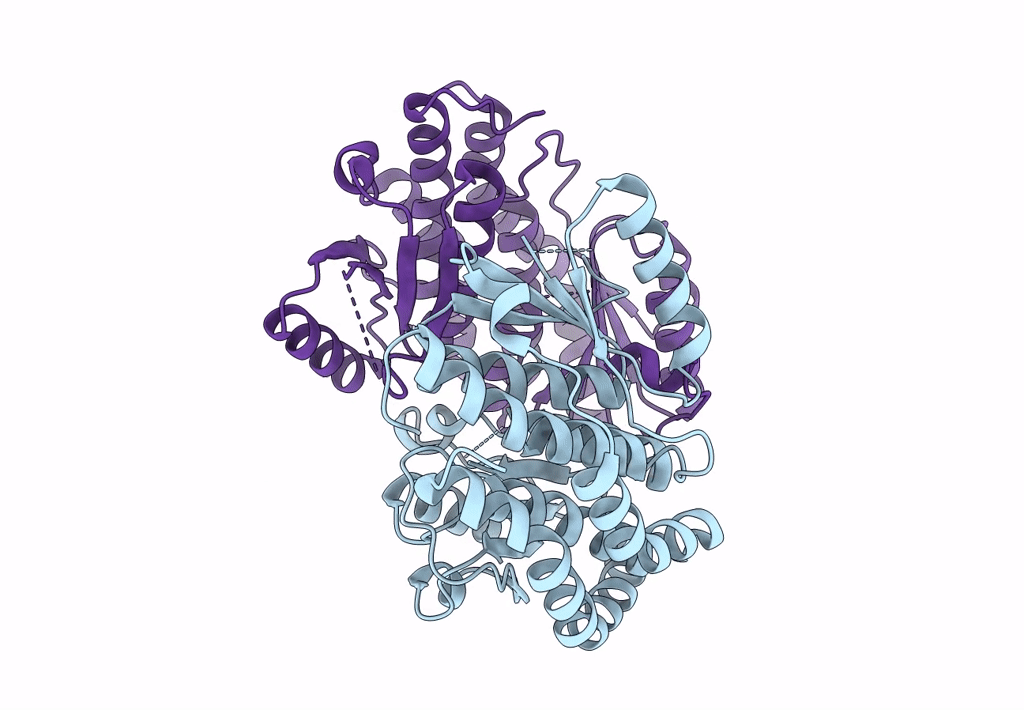
Complex Of Npr1 Ectodomain And Regn5381 Fab In An Active-Like State With No Anp Bound
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: IMMUNE SYSTEM |
 |
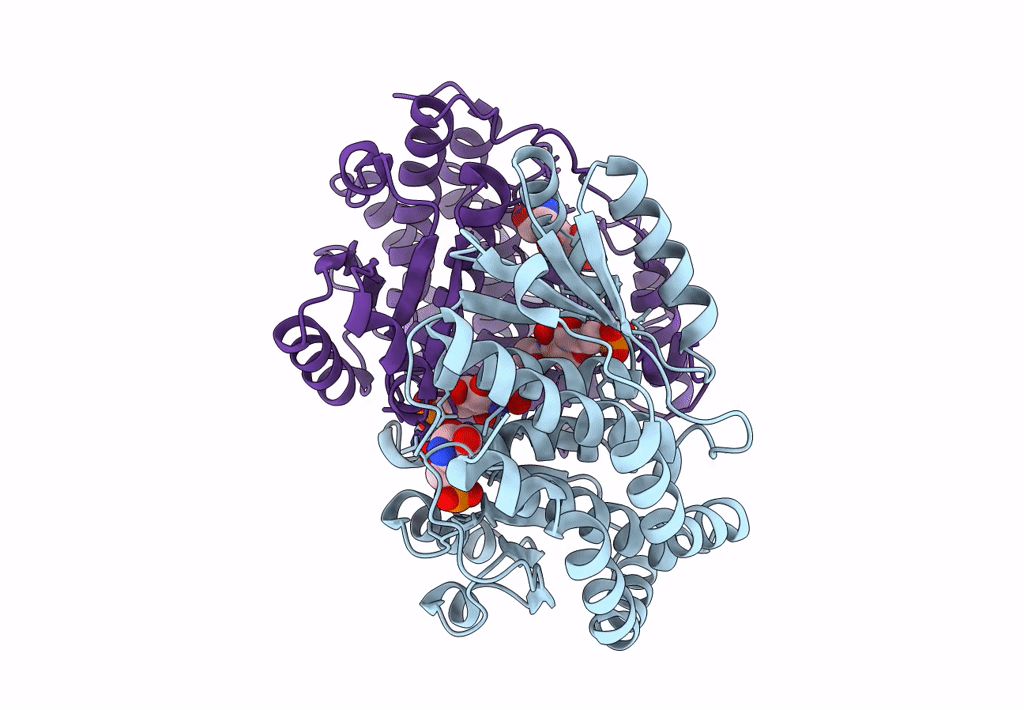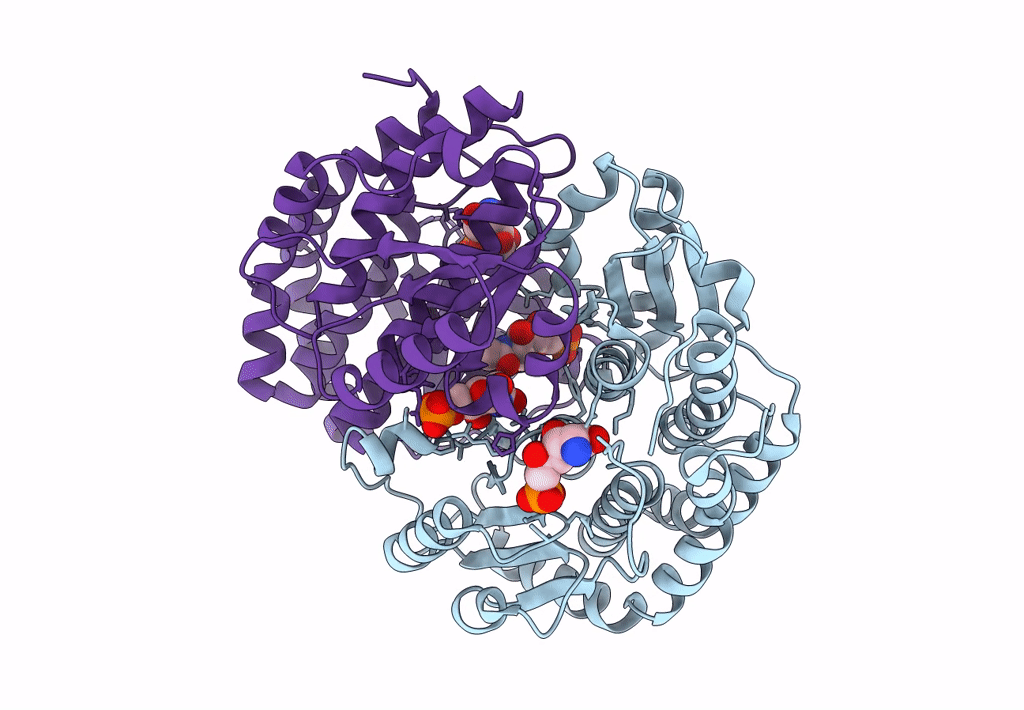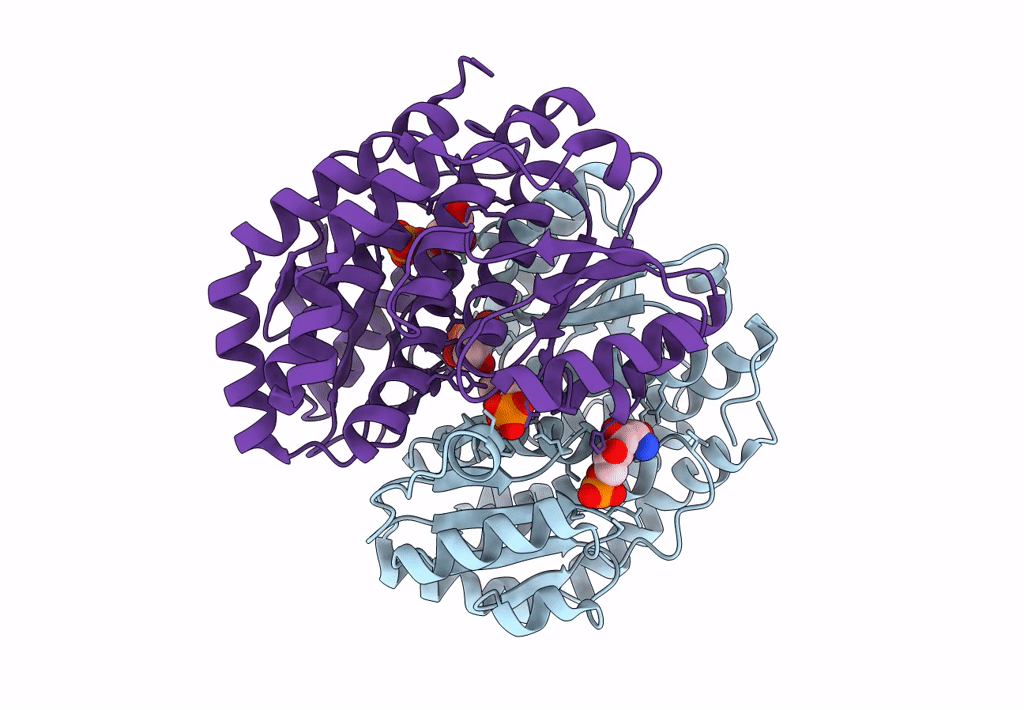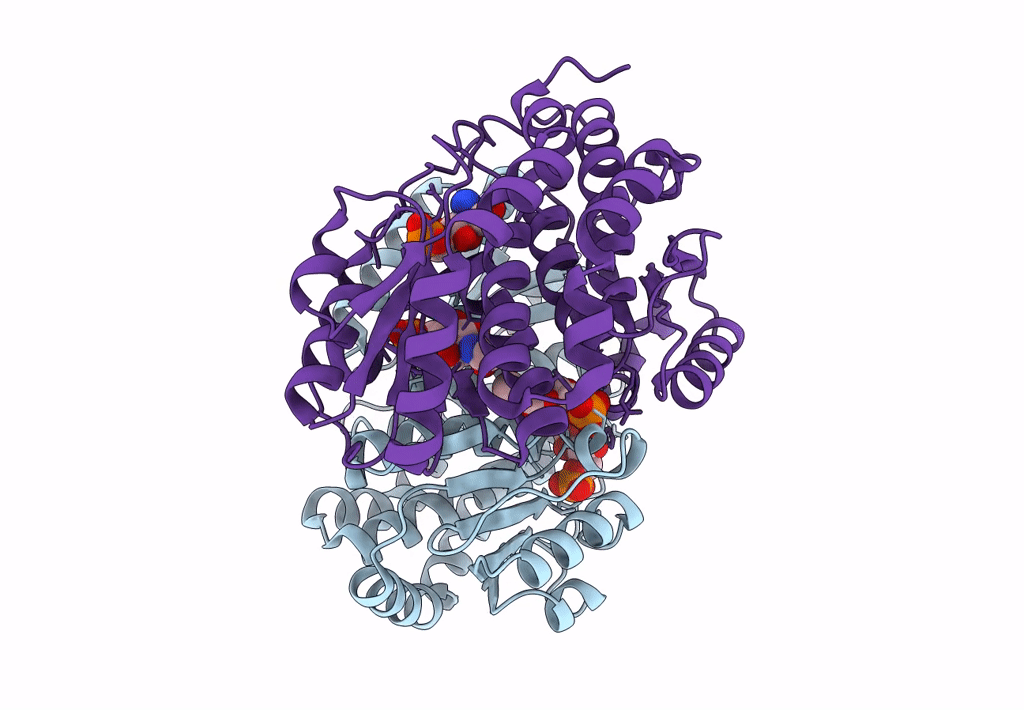
Crystal Structure Of Nagb-Ii Phosphosugar Isomerase From Shewanella Denitrificans Os217 At 2.17 A Resolution
Organism: Shewanella denitrificans os217
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-05-17 Classification: ISOMERASE |
 |
Crystal Structure Of Nagb-Ii Phosphosugar Isomerase From Shewanella Denitrificans Os217 In Complex With Glucitolamine-6-Phosphate And N-Acetylglucosamine-6-Phosphate At 2.31 A Resolution
Organism: Shewanella denitrificans os217
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-05-17 Classification: ISOMERASE Ligands: 16G, AGP |
 |
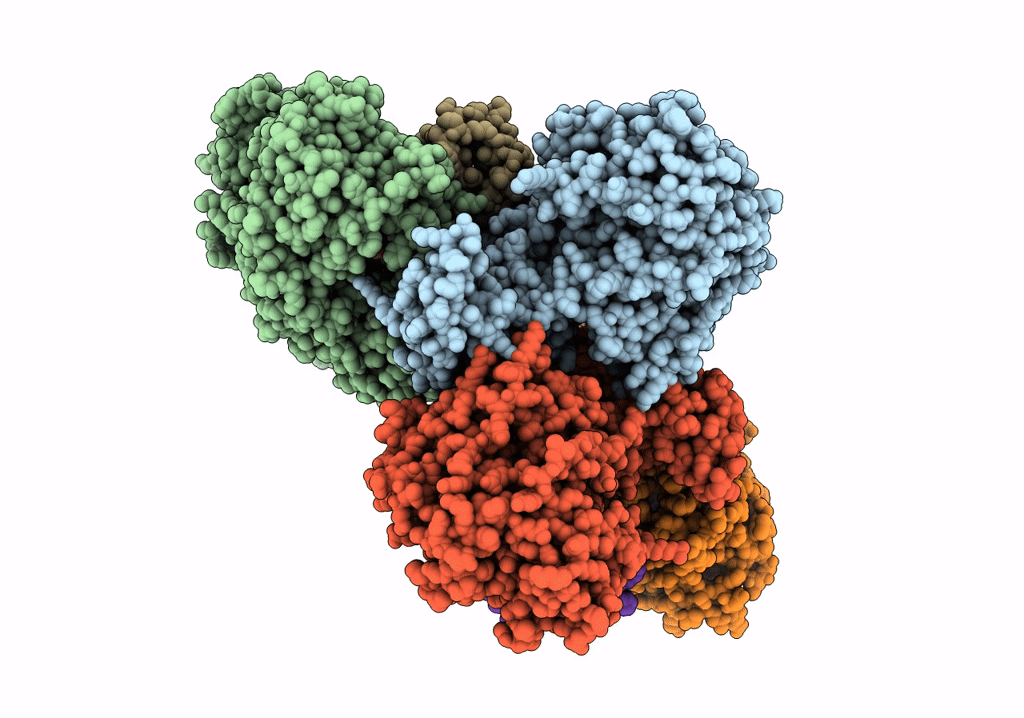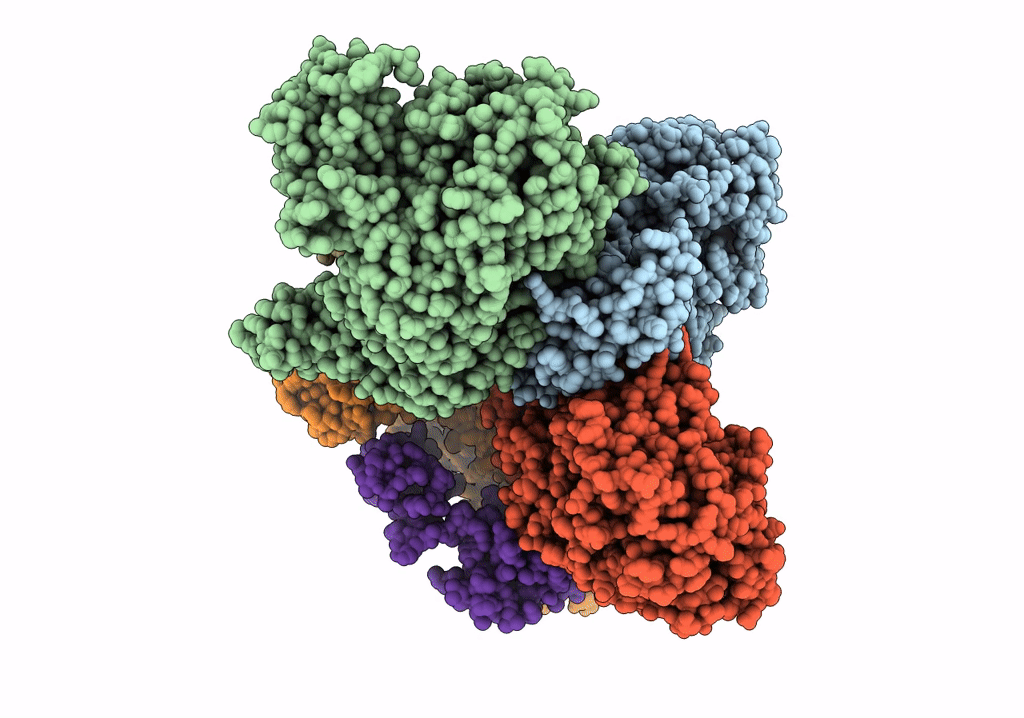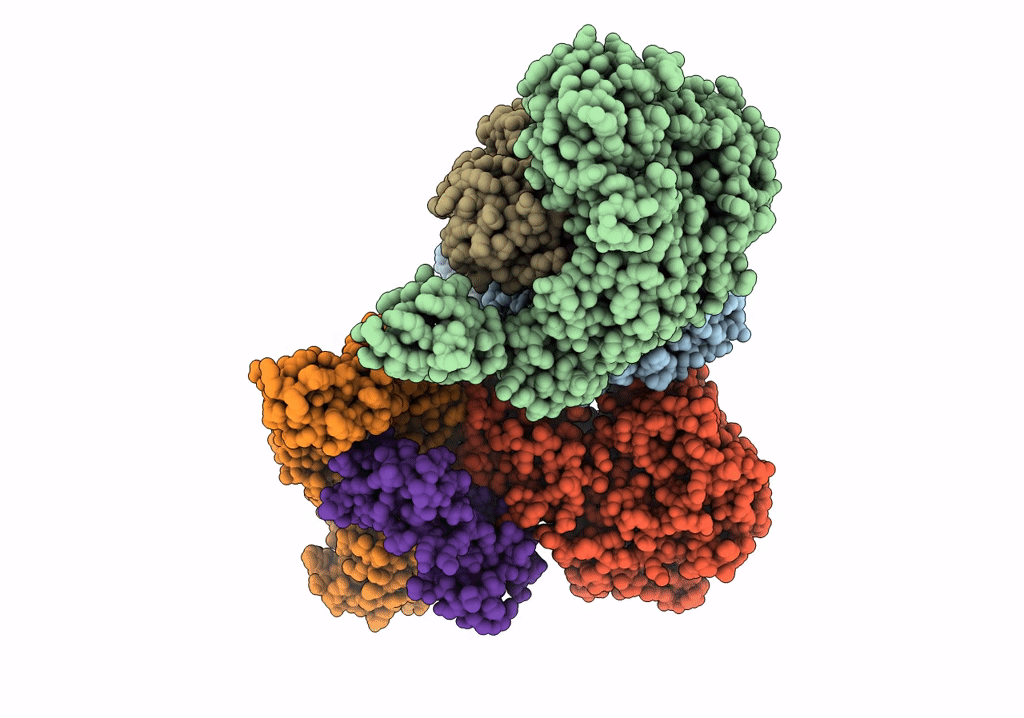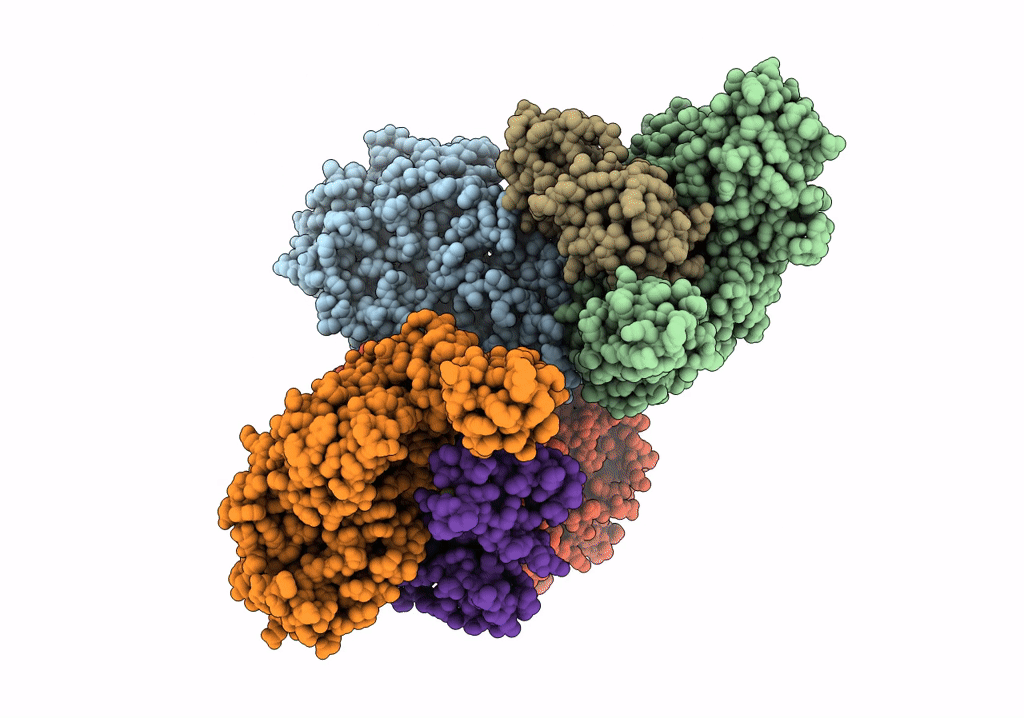
Crystal Structure Of Nagb-Ii Phosphosugar Isomerase From Shewanella Denitrificans Os217 In Complex With Glucitolamine-6-Phosphate At 3.06 A Resolution.
Organism: Shewanella denitrificans os217
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2023-05-17 Classification: ISOMERASE Ligands: AGP, MG, GOL |
 |
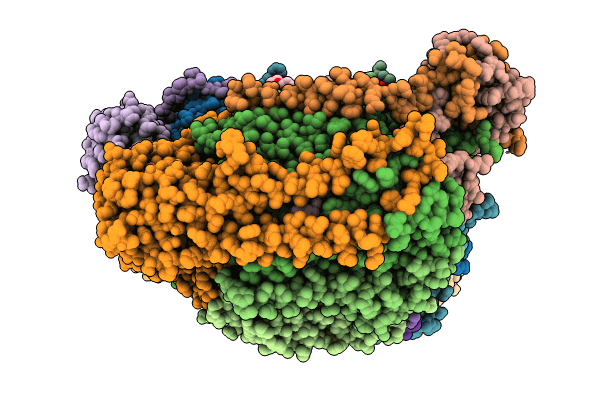
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, ZN, FMN, NAD |
 |
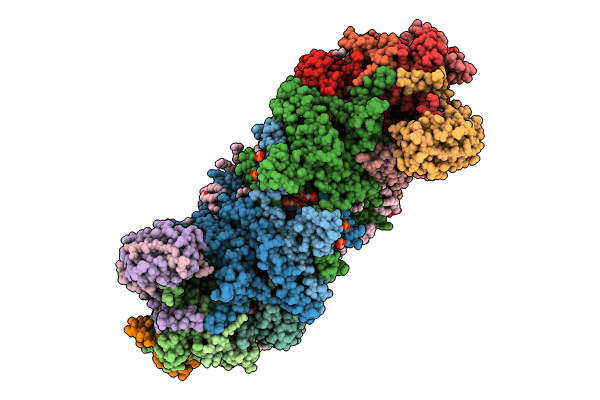
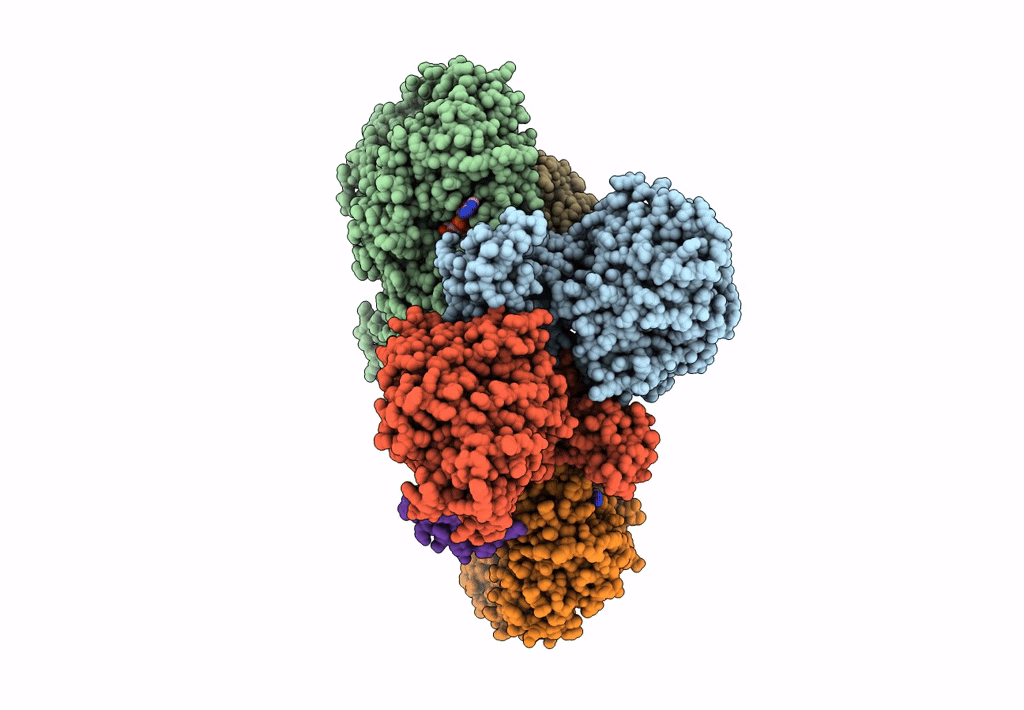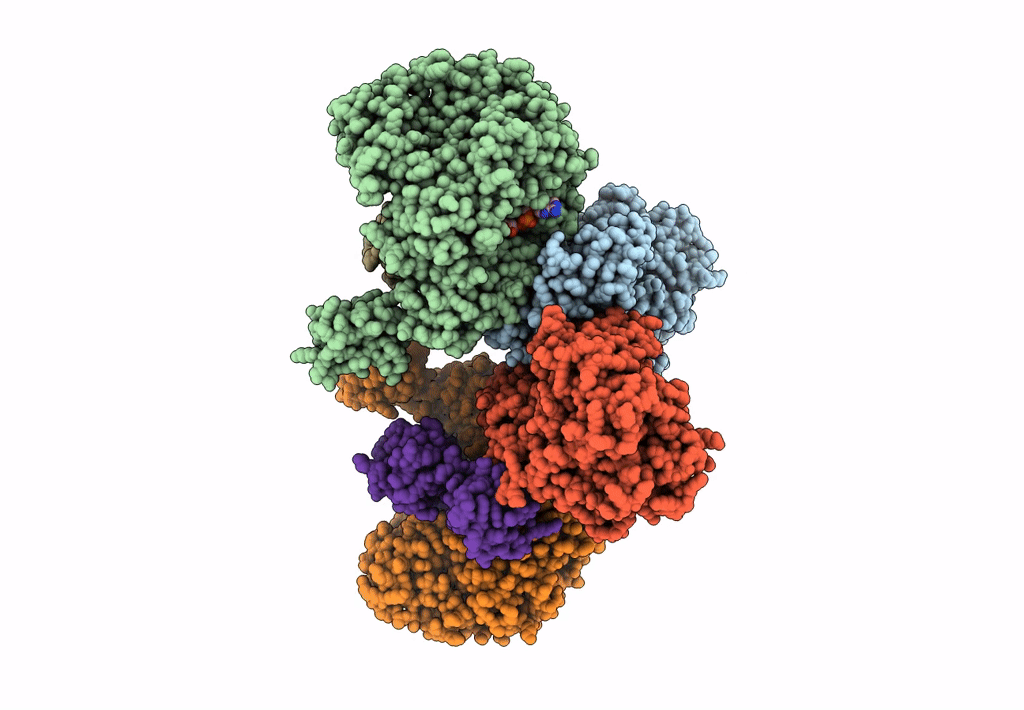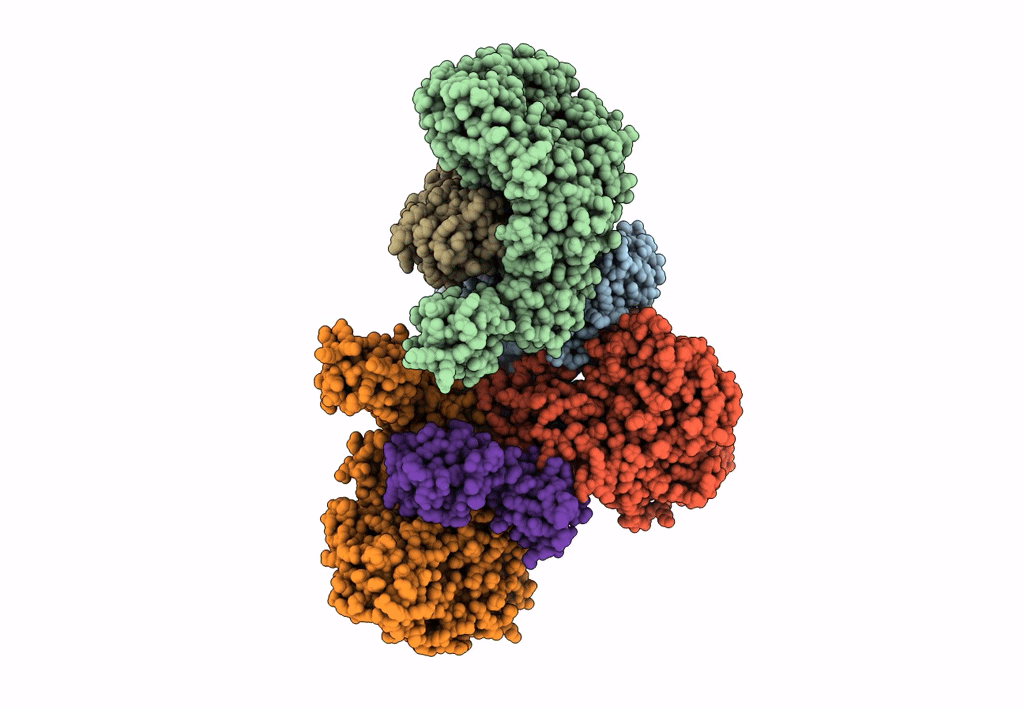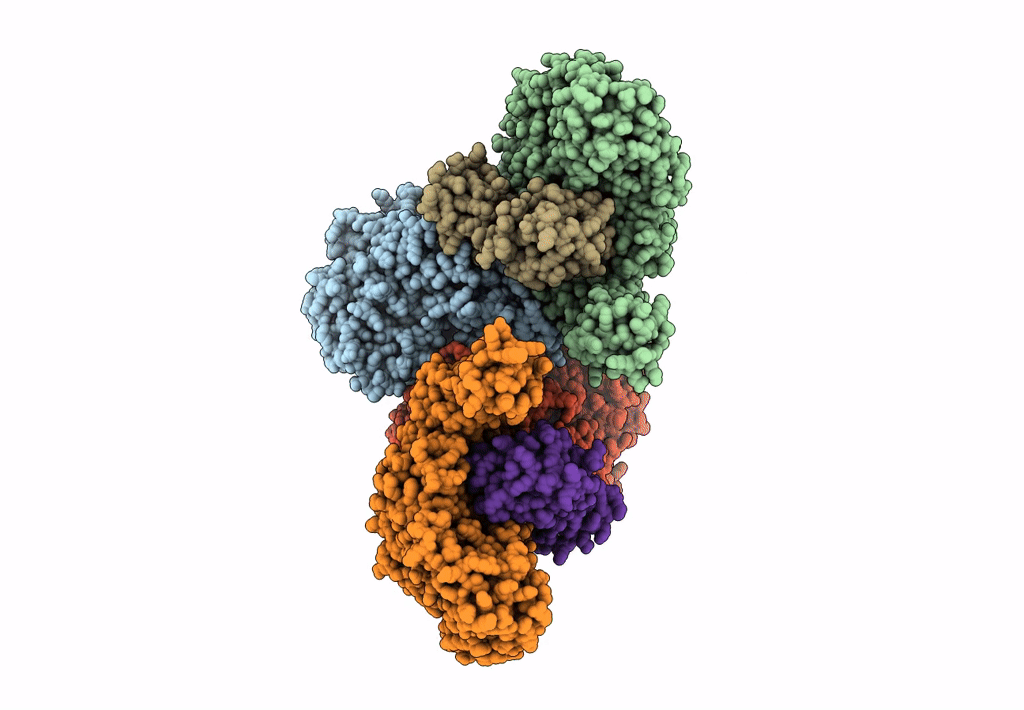
Cryo-Em Structure Of The Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex From Acetobacterium Woodii In The Reduced State
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, HC1, ZN, FMN, NAI |
 |
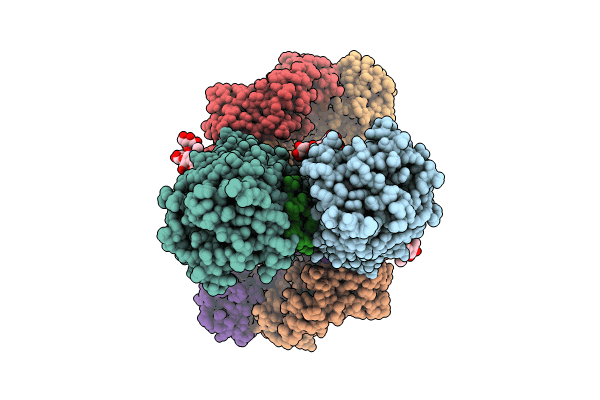
Cryoem Structure Of Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex A. Woodii In The Oxidised State
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FMN, ZN |
 |
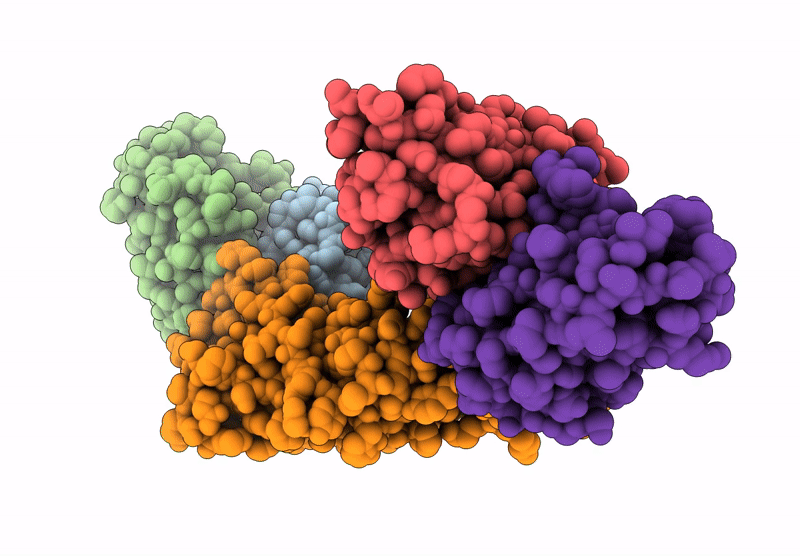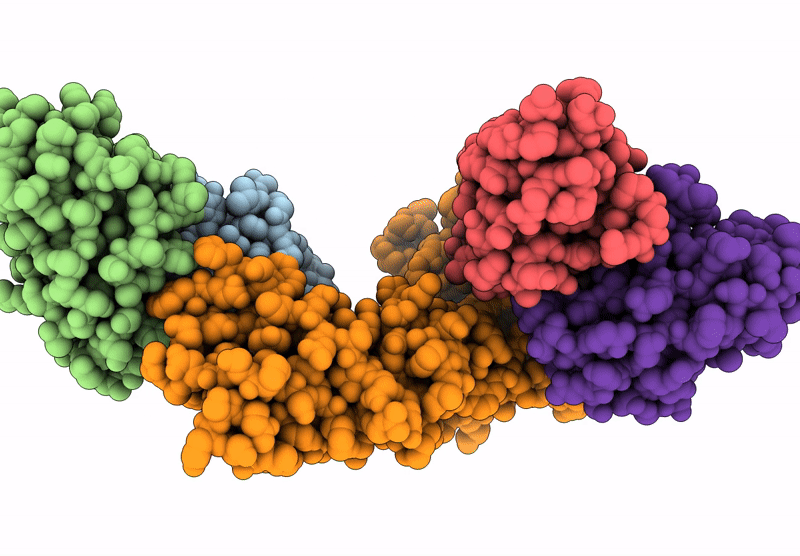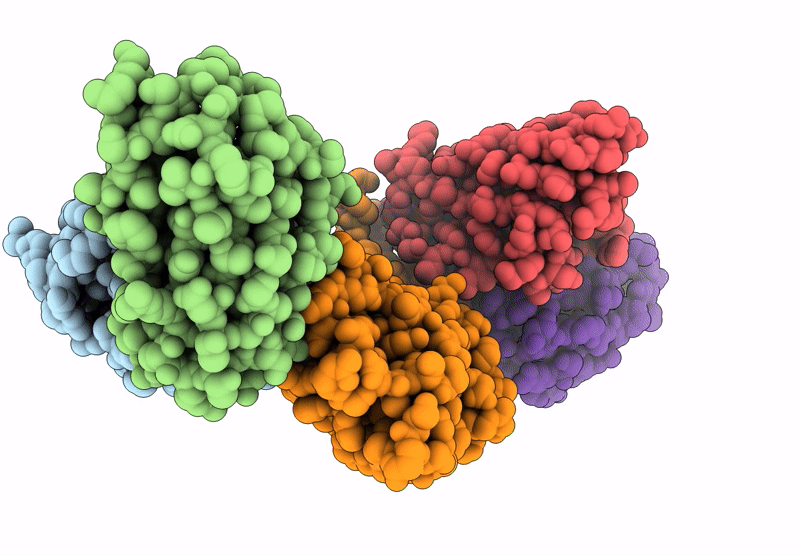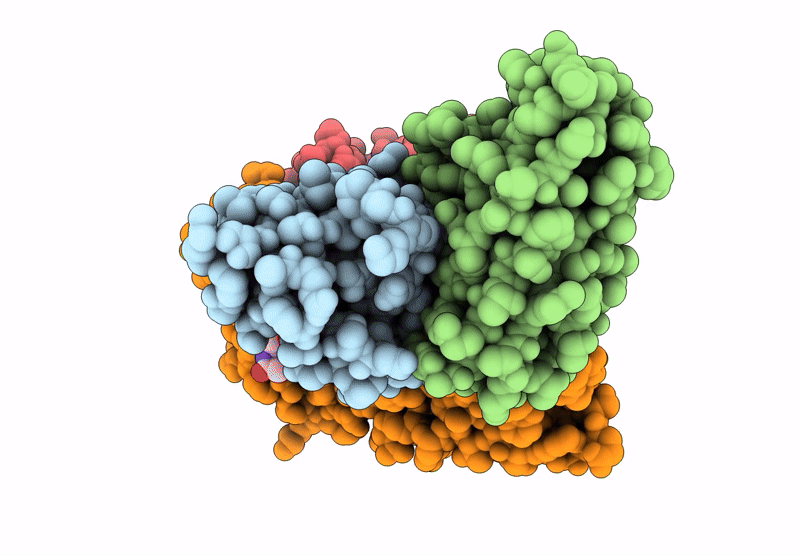
Cryo-Em Structure Of The Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex From Thermoanaerobacter Kivui In The Reduced State
Organism: Thermoanaerobacter kivui
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: SF4, HC1, FES, FMN, ZN, NAP |
 |
Cryo-Em Structure Of The Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex From Thermoanaerobacter Kivui In The Oxidised State
Organism: Thermoanaerobacter kivui
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FMN, ZN |
 |
Structure Of Interleukin Receptor Common Gamma Chain (Il2Rgamma) In Complex With Two Antibodies
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: IMMUNE SYSTEM |

