Search Count: 285
 |
Organism: Streptococcus canis
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Structure Of The Truncated Version Of Idec Protease C94S From Streptococcus Canis
Organism: Streptococcus canis
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
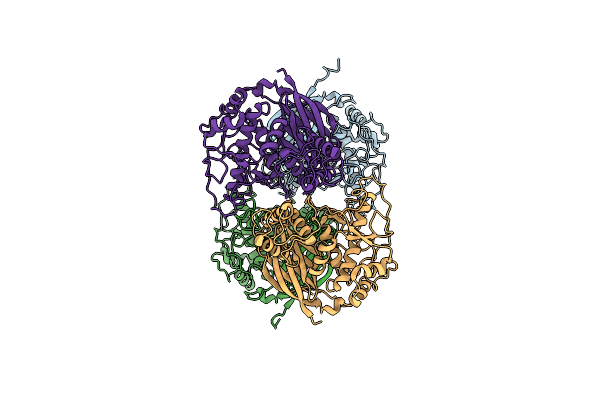 |
Crystal Structure Of The Spore Gernation Lytic Transglycosylase Slec From Clostridioides Difficile In Its Zymogenic Form (Prepro-Slec)
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-12 Classification: HYDROLASE |
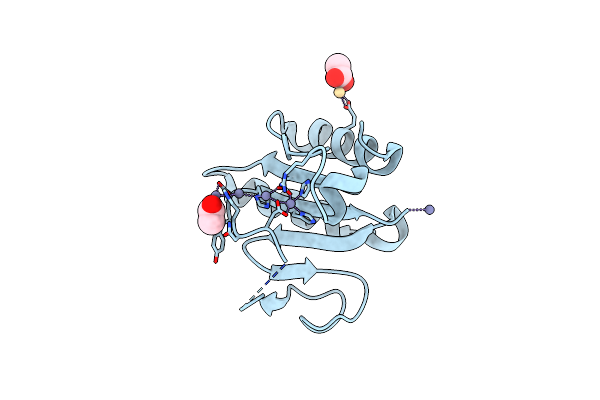 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing Four Zinc Atoms At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CD, ACT |
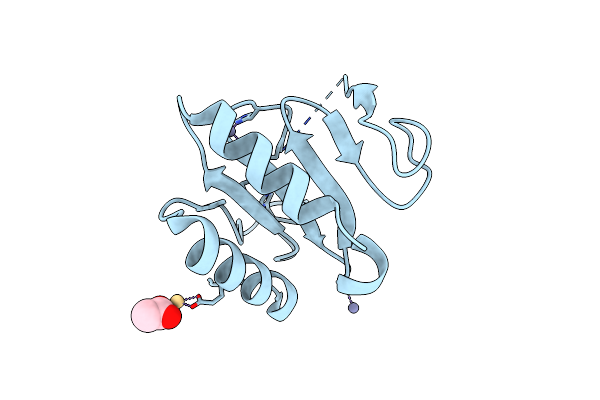 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing Three Zinc Atoms At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ACT, ZN, CD |
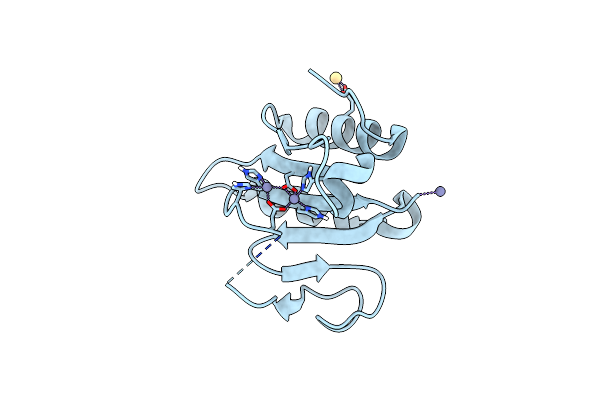 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing Two Zinc Atoms At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CD |
 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing A Zinc Atom At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ACT, CD, ZN |
 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae In A Catalytically Competent Conformation
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The C-Terminal Domain Of Vlde H373A From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of Ami1 From M. Tuberculosis In Complex With A Tetrazole Compound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-11-20 Classification: HYDROLASE Ligands: A1A0I, ZN |
 |
Crystal Structure Of Cyclophilin Tgcyp23 From Toxoplasma Gondii In Complex With Nim811 (N-Methyl-4-Isoleucine Cyclosporin)
Organism: Toxoplasma gondii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2024-10-30 Classification: ISOMERASE |
 |
Crystal Structure Of Cyclophilin Tgcyp23 From Toxoplasma Gondii In Complex With Alisporivir (Nonimmunosuppressive Analogue Of Cyclosporin)
Organism: Toxoplasma gondii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-10-30 Classification: ISOMERASE |
 |
Crystal Structure Of Cyclophilin Tgcyp23 From Toxoplasma Gondii In Complex With Dihydro Cyclosporin A
Organism: Toxoplasma gondii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-10-30 Classification: ISOMERASE |
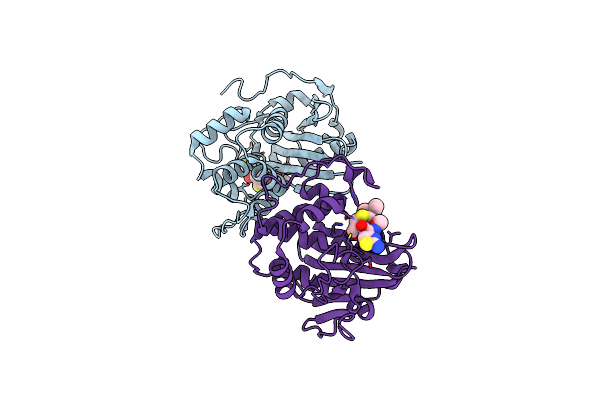 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With Cefepime
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-09-04 Classification: SIGNALING PROTEIN Ligands: UJ9 |
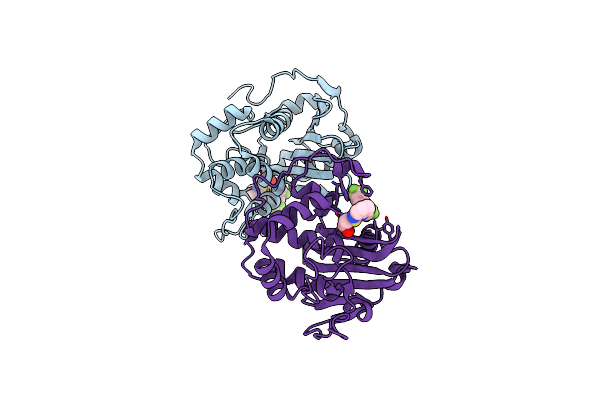 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With A Boronate Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-10 Classification: SIGNALING PROTEIN Ligands: SZI |
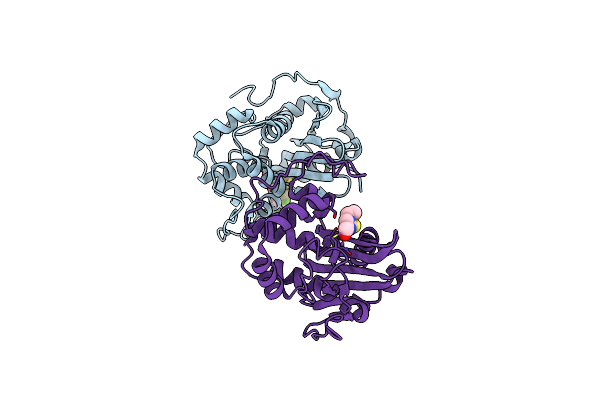 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With An Imidazole Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-07-10 Classification: SIGNALING PROTEIN Ligands: SYU |
 |
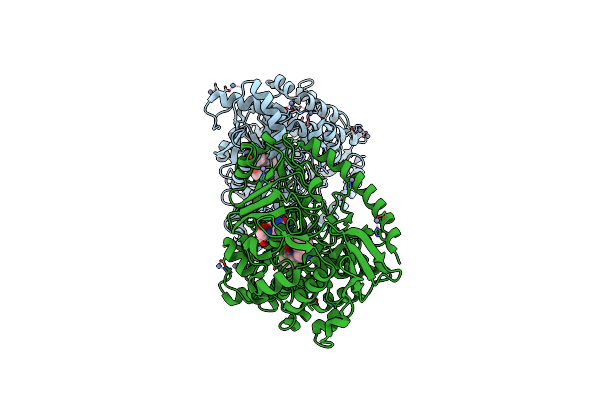
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alid In Open Conformation
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN Ligands: MG |
 |
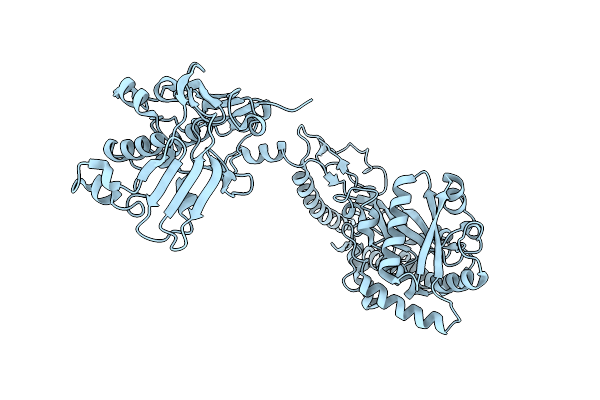
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alid In Closed Conformation In Complex With Peptide 1
Organism: Streptococcus pneumoniae, Prevotella
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
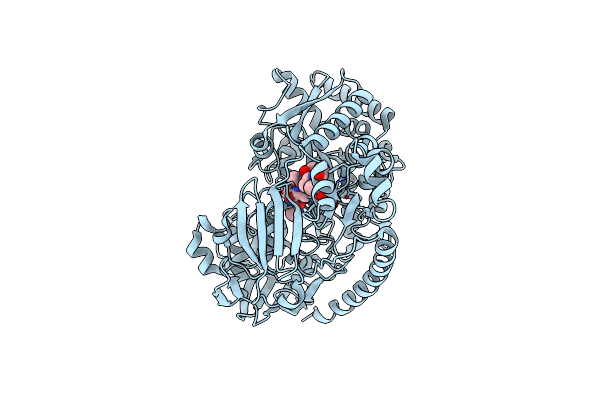
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alic As A Domain-Swapped Dimer
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alib In Complex With An Unknown Peptide
Organism: Streptococcus pneumoniae, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |

