Search Count: 7
 |
Apo Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-05-20 Classification: HYDROLASE |
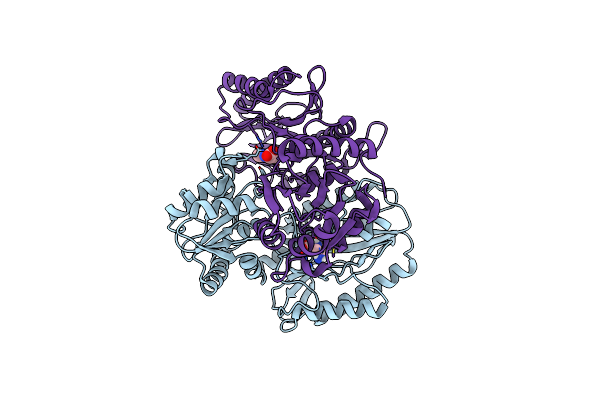 |
Substrate Bound Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: P4B, 4CS |
 |
Apo Structure Of The Ectoine Utilization Protein Eute (Doeb) From Ruegeria Pomeroyi
Organism: Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Product Bound Structure Of The Ectoine Utilization Protein Eute (Doeb) From Ruegeria Pomeroyi
Organism: Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN, DAB, ACT |
 |
Product Bound Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, P4B |
 |
Crystal Structure Of The Beta-Hydroxyaspartate Aldolase Of Paracoccus Denitrificans
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-08-14 Classification: LYASE Ligands: PLP, MG |
 |
Crystal Structure Of The Iminosuccinate Reductase Of Paracoccus Denitrificans In Complex With Nad+
Organism: Paracoccus denitrificans (strain pd 1222)
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2019-08-14 Classification: OXIDOREDUCTASE Ligands: NAD, TB, 7MT |

