Search Count: 18
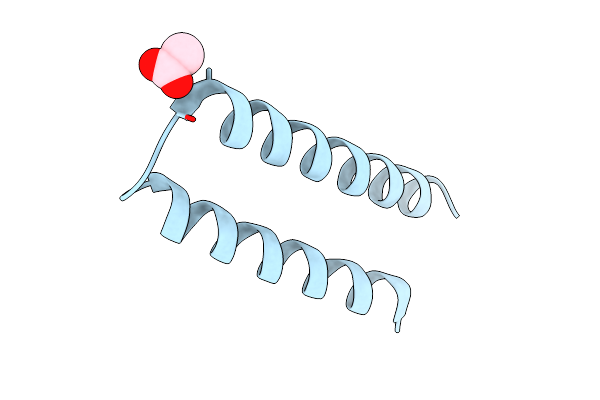 |
X-Ray Crystal Structure Of A De Novo Designed Helix-Loop-Helix Homodimer In An Anti Arrangement, Cc-Hp1.0
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-06-07 Classification: DE NOVO PROTEIN Ligands: ACT |
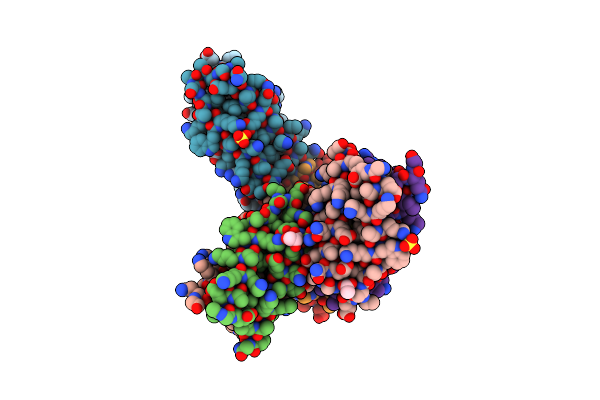 |
X-Ray Crystal Structure Of A De Novo Selected Helix-Loop-Helix Heterodimer In A Syn Arrangement, 26Alpha/26Beta
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-07 Classification: DE NOVO PROTEIN Ligands: SO4, GOL, ACT |
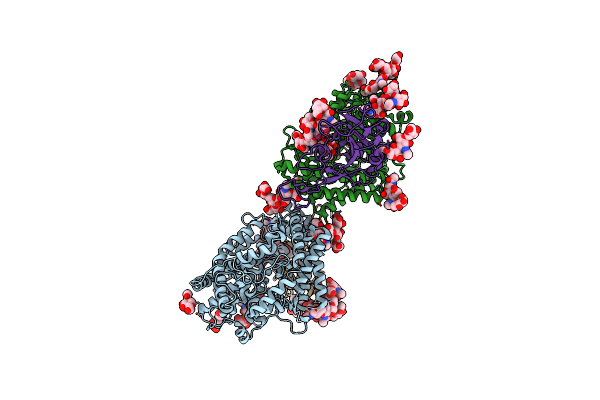 |
Crystal Structure Of Spike Protein Receptor Binding Domain Of Escape Mutant Sars-Cov-2 From Immunocompromised Patient (D146*) In Complex With Human Receptor Ace2
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2021-12-08 Classification: VIRAL PROTEIN Ligands: ZN, CL, NAG |
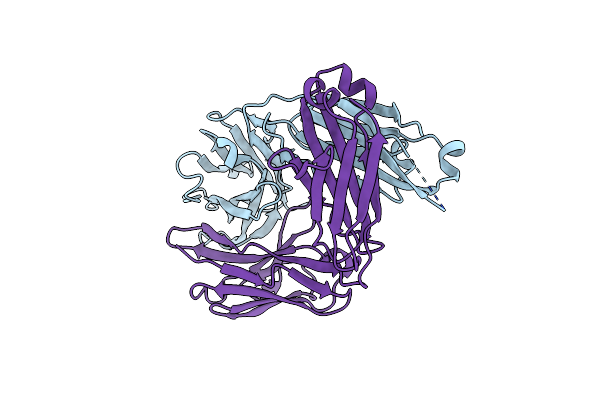 |
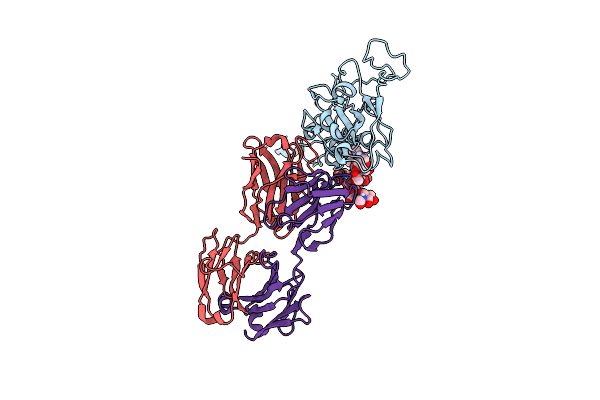
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2021-12-08 Classification: IMMUNE SYSTEM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: IMMUNE SYSTEM |
 |
Structure Of Human Sars-Cov-2 Spike Glycoprotein Trimer Bound By Neutralizing Antibody C1C-A3 Fab (Variable Region)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-11-14 Classification: IMMUNE SYSTEM |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp.r39
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-02-29 Classification: HYDROLASE Ligands: B07, SO4, MG |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Release Date: 2012-02-29 Classification: HYDROLASE Ligands: SO4, MG, FP5, ACN, 33D, MES |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-07-27 Classification: HYDROLASE Ligands: BH6, SO4, MG |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Release Date: 2011-07-27 Classification: HYDROLASE Ligands: FP5, SO4, MG, ACN |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-07-27 Classification: HYDROLASE Ligands: ZA3, SO4, MG |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-08-18 Classification: LYASE Ligands: ZN, SO4, BCT |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2009-06-02 Classification: LYASE Ligands: ZN |
 |
H. Influenzae Beta-Carbonic Anhydrase, Variant D44N In 100 Mm Sodium Bicarbonate
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-06-02 Classification: LYASE Ligands: ZN |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-06-02 Classification: LYASE Ligands: ZN, PO4 |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-06-02 Classification: LYASE Ligands: ZN, SO4 |
 |
H. Influenzae Beta-Carbonic Anhydrase, Variant Y181F With 100 Mm Bicarbonate
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-06-02 Classification: LYASE Ligands: ZN, SO4, BCT |

