Search Count: 26
 |
Staphylococcus Aureus 70S Ribosome With Elongation Factor G Locked With Fusidic Acid Cyclopentane In Post-Translocational State
Organism: Staphylococcus aureus subsp. aureus nctc 8325, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RIBOSOME Ligands: ZN, MG, SPD, PUT, GDP, WUX, K |
 |
Staphylococcus Aureus 70S Ribosome With Elongation Factor G Locked With Fusidic Acid Cyclopentane With A Trna In Pe/E Chimeric State
Organism: Staphylococcus aureus subsp. aureus nctc 8325, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RIBOSOME Ligands: MG, SPD, PUT, GDP, WUX, ZN |
 |
Staphylococcus Aureus 70S Ribosome With Elongation Factor G Locked With Fusidic Acid With A Trna In Pe/E Chimeric State
Organism: Staphylococcus aureus subsp. aureus nctc 8325, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RIBOSOME Ligands: MG, SPD, PUT, GDP, FUA, ZN |
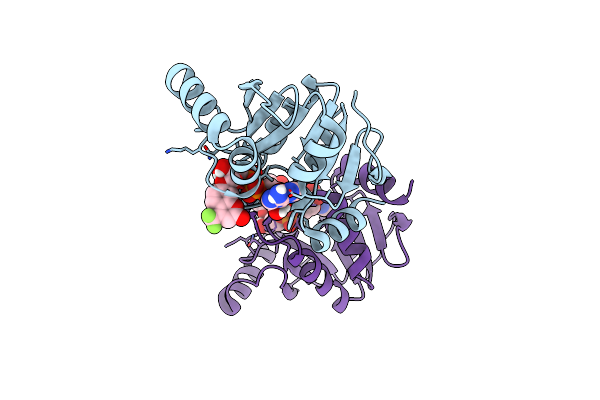 |
Crystal Structure Of Sars-Cov-2 (Covid-19) Nsp3 Macrodomain In Complex With Tfmu-Adpr
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-05-17 Classification: VIRAL PROTEIN Ligands: ZJ3 |
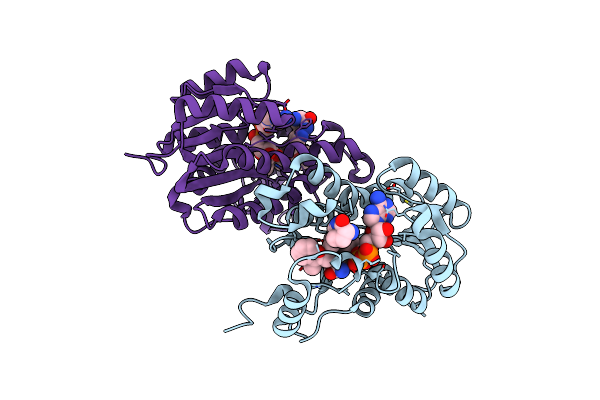 |
Crystal Structure Of E. Coli Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NQF |
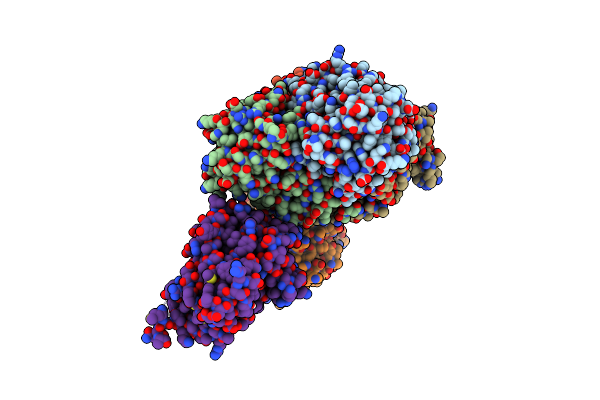 |
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NQF, NAD |
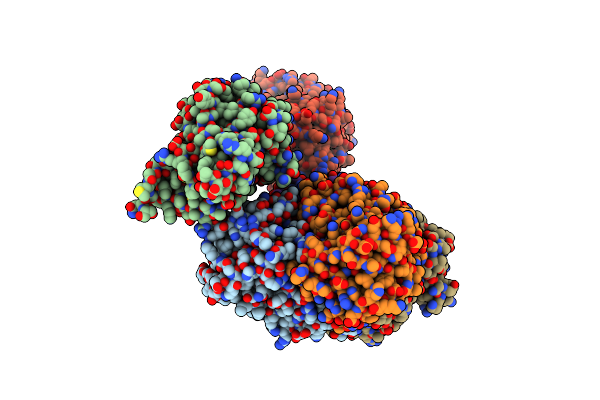 |
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NUC |
 |
Crystal Structure Of E. Coli Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-14 Classification: ANTIBIOTIC Ligands: NAD, NUC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-01-09 Classification: HYDROLASE Ligands: C7G, MG, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: APR, MG, ACY, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2018-12-19 Classification: HYDROLASE |
 |
Organism: Latimeria chalumnae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: MG, CIT, ACT, GOL |
 |
Adp-Ribosylserine Hydrolase Arh3 Of Latimeria Chalumnae In Complex With Adp-Ribose
Organism: Latimeria chalumnae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: MG, AR6 |
 |
Human [Protein Adp-Ribosylargenine] Hydrolase Arh1 In Complex With Adp-Ribose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: AR6, MG, CL |
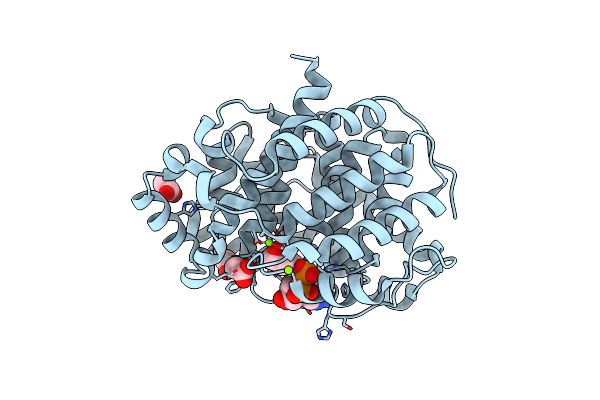 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: A3R, MG, CL, ACT |
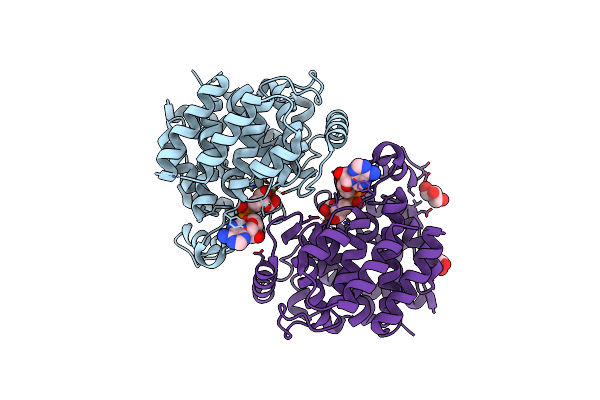 |
Adp-Ribosylserine Hydrolase Arh3 Of Latimeria Chalumnae In Complex With Adp-Ribose
Organism: Latimeria chalumnae
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: MG, AR6, GOL |
 |
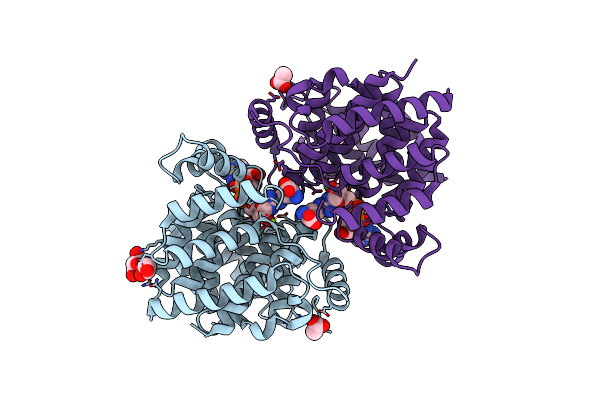
Adp-Ribosylserine Hydrolase Arh3 Of Latimeria Chalumnae In Complex With Adp-Hpd
Organism: Latimeria chalumnae
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: MG, A1R, GOL |
 |
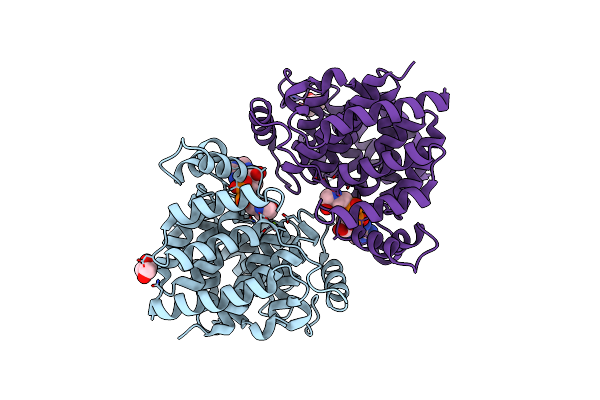
Adp-Ribosylserine Hydrolase Arh3 Of Latimeria Chalumnae In Complex With Adp-Ribosyl-L-Arginine
Organism: Latimeria chalumnae
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: MG, F2R, GOL, ACT |
 |
Adp-Ribosylserine Hydrolase Arh3 Of Latimeria Chalumnae In Complex With Adp-Hpm
Organism: Latimeria chalumnae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: MG, A3R, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-11-28 Classification: HYDROLASE Ligands: A3R, SO4 |

