Search Count: 44
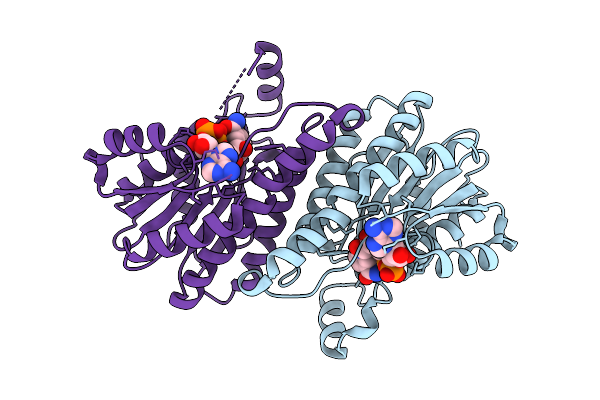 |
Crystal Structure Of L-2-Keto-3-Deoxypentonate 4-Dehydrogenase Bound To Nad(H)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crysral Structure Of 2-Keto-3-Deoxypentonate 4-Dehydrogenase From Herbaspirillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase From Herbaspillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: PEG |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To Nad(H) And Sulfate Ion
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, PEG, TRS, GOL |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-Kdf Or L-2,4-Dkdf
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXA, A1LXB, PEG |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-2,4-Dkdf And Nadh
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, A1LXB |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To D-Kdp
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXD, PEG |
 |
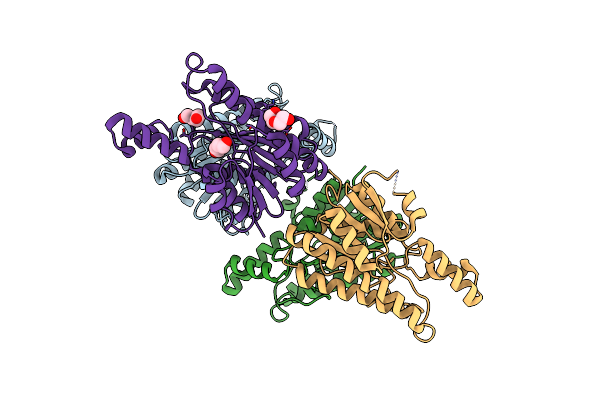
Crystal Structure Of Hydroxyisourate Hydrolase From Herbaspirillum Seropedicae
Organism: Herbaspirillum seropedicae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-01-31 Classification: HYDROLASE |
 |
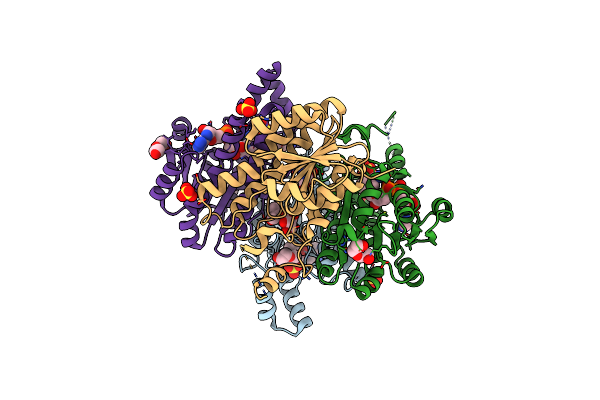
Crystal Structure Of Cf, A Heterohexamer Of The 4-Oxalocrotonate Tautomerase (4-Ot) Family
Organism: Herbaspirillum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-06 Classification: ISOMERASE |
 |
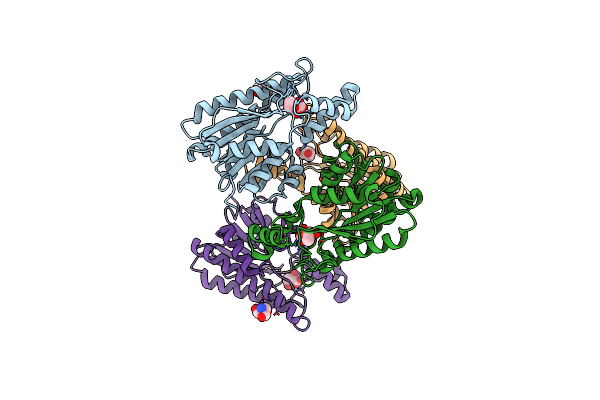
Crystal Structure Of Yr, A Heterohexamer Of The 4-Oxalocrotonate Tautomerase (4-Ot) Family
Organism: Herbaspirillum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Organism: Herbaspirillum
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Crystal Structure Of Herbaspirillum Huttiense L-Arabinose 1-Dehydrogenase (Nad Bound Form)
Organism: Herbaspirillum huttiense subsp. putei iam 15032
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE Ligands: NAD, PEG |
 |
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: BME |
 |
Triuret Hydrolase (Trta) From Herbaspirillum Sp. Bh-1 C162S Bound With Biuret
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: EDO, C5J |
 |
Triuret Hydrolase (Trta) From Herbaspirillum Sp. Bh-1 C162S Bound With Triuret
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: TIU, EDO |
 |
Organism: Herbaspirillum seropedicae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-08-03 Classification: DNA BINDING PROTEIN Ligands: ADP, ATP, CA |
 |
Crystal Structure Of The Rna Chaperone Hfq From Herbaspirillum Seropedicae Smr1
Organism: Herbaspirillum seropedicae
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2012-01-04 Classification: CHAPERONE Ligands: GOL |
 |
Organism: Herbaspirillum seropedicae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-06-17 Classification: SIGNALING PROTEIN |
 |
Organism: Noviherbaspirillum sp. Root189
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Noviherbaspirillum sp. Root189
Method: Alphafold Release Date: Classification: NA Ligands: NA |

