Search Count: 19
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: CL, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: CL, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: CL, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: CL, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-03-05 Classification: SIGNALING PROTEIN Ligands: PO4, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-22 Classification: SIGNALING PROTEIN Ligands: CYS, NA, CL |
 |
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-12-28 Classification: DNA BINDING PROTEIN |
 |
Organism: Geobacillus stearothermophilus atcc 7953
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2022-12-28 Classification: DNA BINDING PROTEIN |
 |
Structure Of An Intellectual Disability-Associated Ornithine Decarboxylase Variant G84R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-10-19 Classification: HYDROLASE Ligands: PO4 |
 |
Structure Of An Intellectual Disability-Associated Ornithine Decarboxylase Variant G84R In Complex With Plp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-19 Classification: HYDROLASE Ligands: PLP |
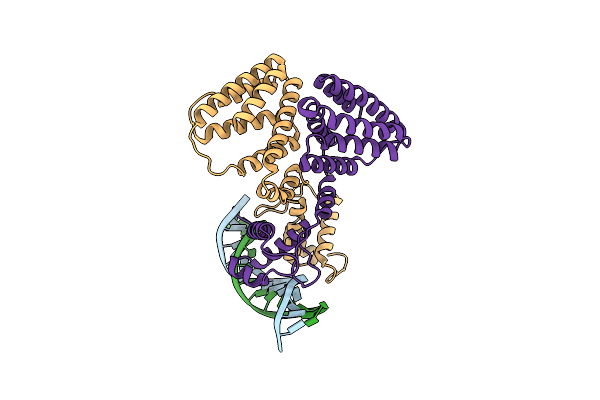 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: GENE REGULATION |
 |
Coordinates Of Nanr Dimer Fitted In Hexameric Nanr-Dna Hetero-Complex Cryo-Em Map
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: GENE REGULATION |

