Search Count: 15
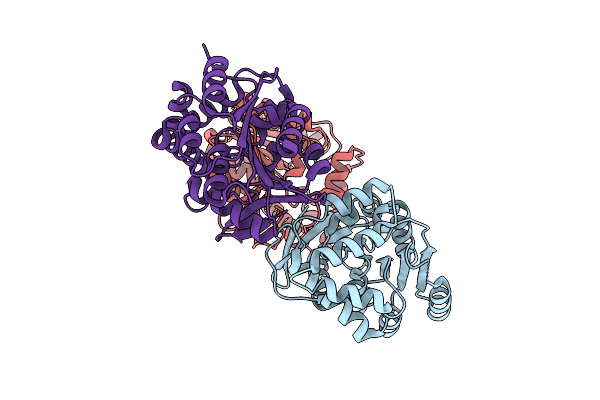 |
Structural Basis For Recognition And Ring-Cleavage Of The Pseudomonas Quinolone Signal (Pqs) By Aqdc
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-03 Classification: HYDROLASE |
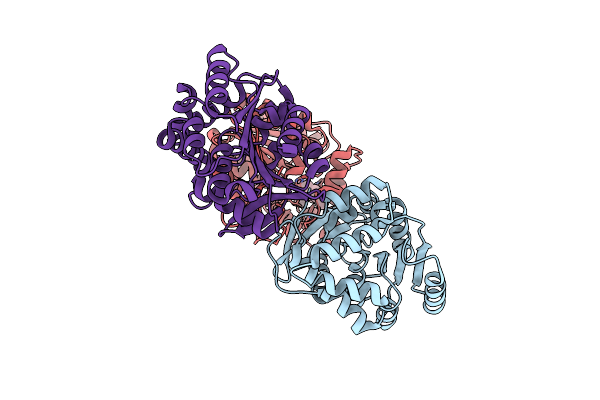 |
Structural Basis For Recognition And Ring-Cleavage Of The Pseudomonas Quinolone Signal (Pqs) By Aqdc In Complex With Its Product
Organism: Mycobacteroides abscessus subsp. abscessus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JWH |
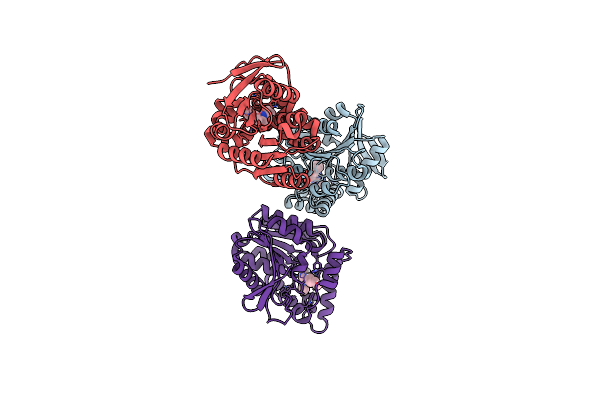 |
Structural Basis For Recognition And Ring-Cleavage Of The Pseudomonas Quinolone Signal (Pqs) By Aqdc Variant In Complex With Its Substrate
Organism: Mycobacterium abscessus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JWW |
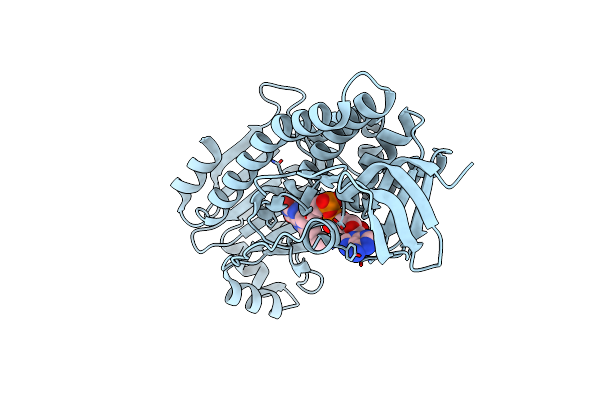 |
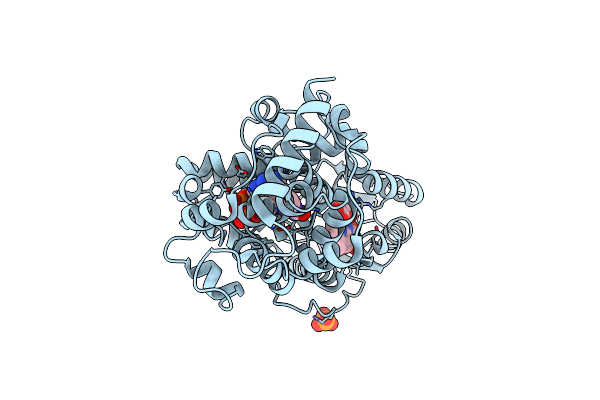
Crystal Structure Of Pqsl, A Probable Fad-Dependent Monooxygenase From Pseudomonas Aeruginosa - New Refinement
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: FAD |
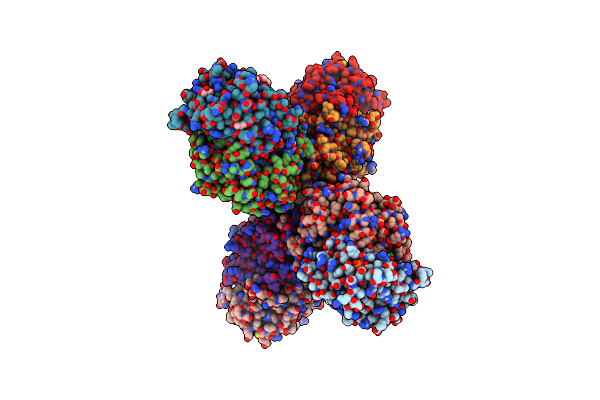 |
Structural And Functional Characterization Of Pqsbc, A Condensing Enzyme In The Biosynthesis Of The Pseudomonas Aeruginosa Quinolone Signal
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: PO4, GOL, MPO, TRS, CL, PGE, PEG, NA |
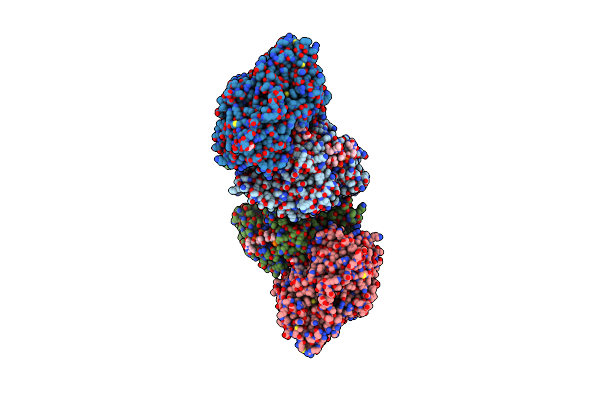 |
Organism: Arabidopsis thaliana, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2009-06-02 Classification: LYASE/DNA Ligands: FAD, MHF, CL, ACT |
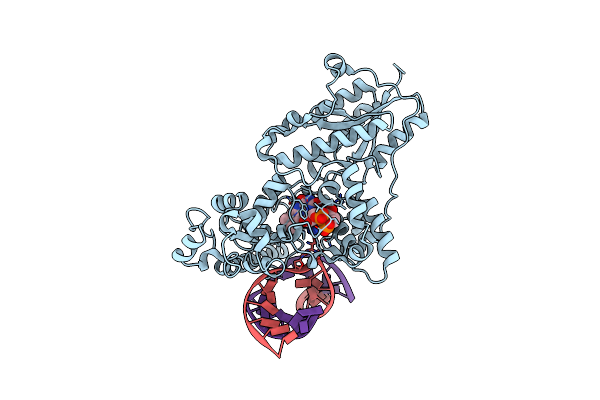 |
Drosophila Melanogaster (6-4) Photolyase Bound To Double Stranded Dna Containing A T(6-4)C Photolesion
Organism: Drosophila melanogaster, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2009-03-17 Classification: LYASE/DNA Ligands: FAD |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2007-05-01 Classification: LYASE Ligands: FAD, HDF, CL, PO4 |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2007-05-01 Classification: LYASE Ligands: CL, FAD, IRF, PO4 |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-05-01 Classification: LYASE Ligands: CL, FAD, FMN, PO4 |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2007-02-20 Classification: TRANSFERASE Ligands: MG, ZN |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2007-02-20 Classification: TRANSFERASE Ligands: MG, ZN |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2007-02-20 Classification: TRANSFERASE Ligands: MG, ZN |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2007-02-20 Classification: TRANSFERASE Ligands: MG, ZN |
 |
Organism: Synechococcus elongatus pcc 6301, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-12-14 Classification: LYASE/DNA Ligands: MG, FAD, HDF |

