Search Count: 101
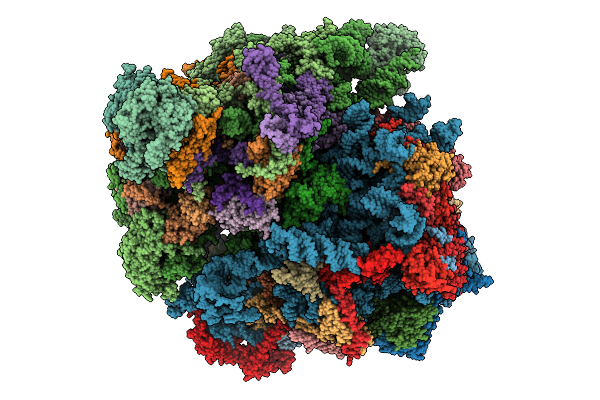 |
Mitochondrial Ribosome Of Saccharomyces Cerevisiae Class Ii From Yep With Dextrose Culture
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: RIBOSOME |
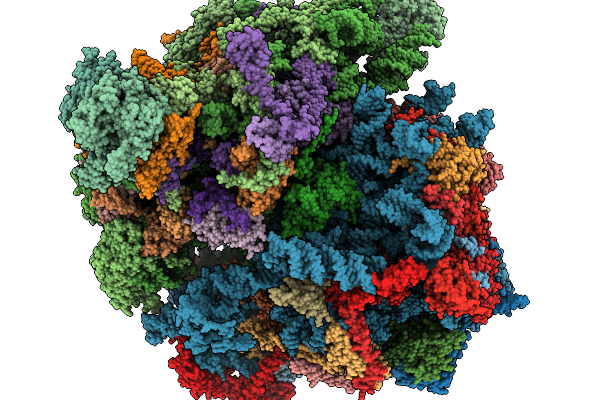 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-08-27 Classification: DNA BINDING PROTEIN |
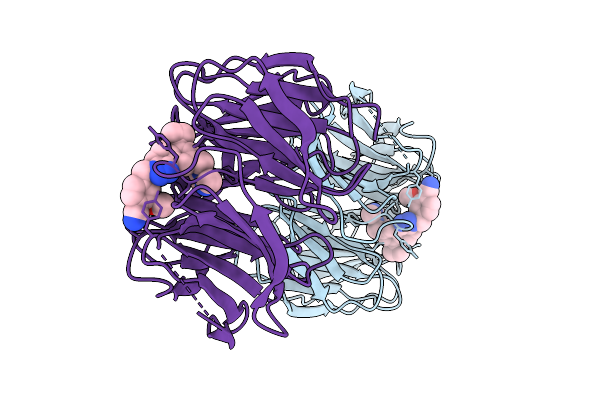 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW7 |
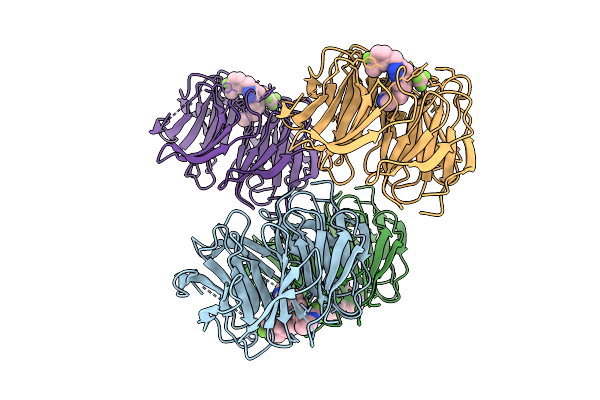 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW9 |
 |
Crystal Structure Of A Beta-1,4-Endoglucanase From Bispora Sp. Mey-1 In Complex With Mannotetrose
Organism: Bispora sp. mey-1
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: NAG, PEG |
 |
Organism: Lysinibacillus composti
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-04-09 Classification: OXIDOREDUCTASE Ligands: EDO, NAD |
 |
Organism: Poliovirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRAL PROTEIN Ligands: SPH |
 |
Organism: Poliovirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-07-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1AWT |
 |
Structure Of The Saccharomyces Cerevisiae Pcna Clamp Unloader Elg1-Rfc Complex
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: REPLICATION Ligands: AGS, MG, ADP |
 |
Structure Of The Saccharomyces Cerevisiae Clamp Unloader Elg1-Rfc Bound To A Cracked Pcna
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: REPLICATION Ligands: AGS, MG, ADP |
 |
Structure Of The Saccharomyces Cerevisiae Clamp Unloader Elg1-Rfc Bound To Pcna
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: REPLICATION Ligands: AGS, MG, ADP |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Sars-Cov-2 S Ntd (C.37 Lambda Variant) Plus S2L20 And S2X303 Fabs, Local Refinement
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-03-13 Classification: ISOMERASE |
 |
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / jcm 6841 / ccug 19606 / cip 70.34 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-03-13 Classification: DNA BINDING PROTEIN |
 |
Organism: Lysinibacillus composti
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-02-14 Classification: OXIDOREDUCTASE Ligands: EDO |

