Search Count: 25
 |
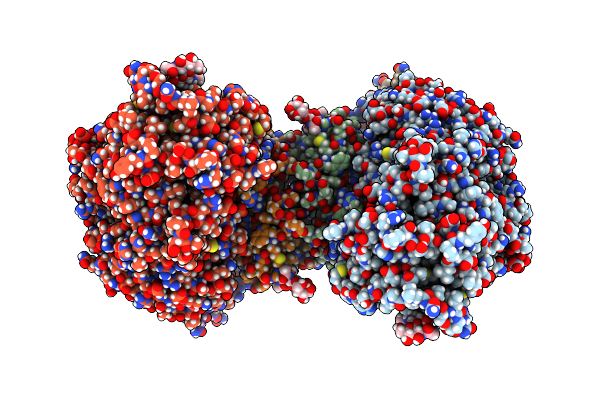
Periplasmic Scaffold Of The Campylobacter Jejuni Flagellar Motor (Alpha Carbon Trace)
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: STRUCTURAL PROTEIN |
 |
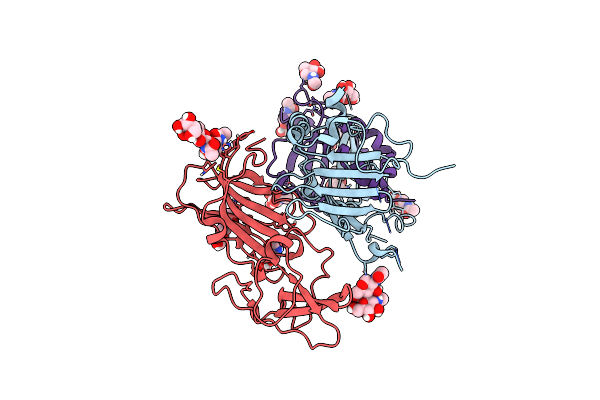
Organism: Human betaherpesvirus 5
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human betaherpesvirus 5
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: VIRAL PROTEIN Ligands: NAG |
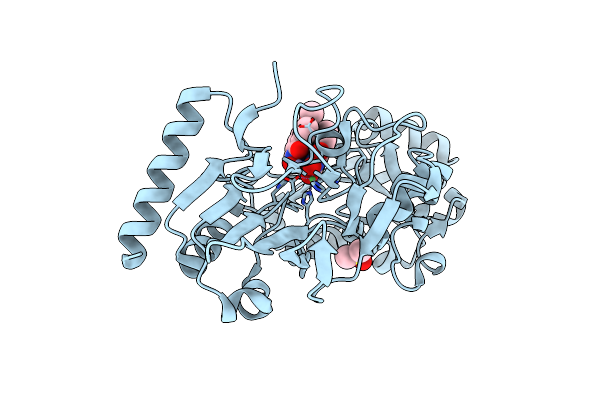 |
Crystal Structure Of Human Dna Cross-Link Repair 1A In Complex With N-Hydroxyimide Inhibitor H1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: DMS, NI, ZN, UFI |
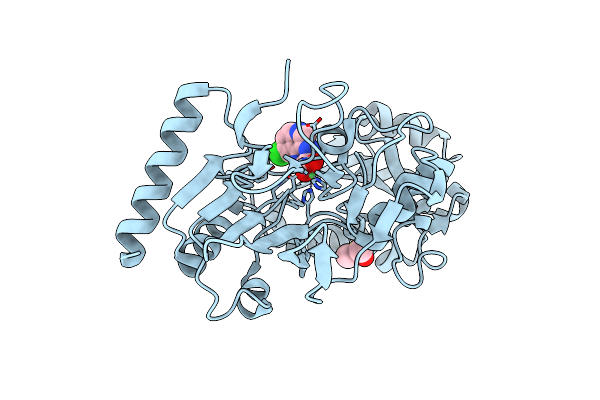 |
Crystal Structure Of Human Dna Cross-Link Repair 1A In Complex With Quinoxalinedione Inhibitor H2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: DMS, UF3, NI |
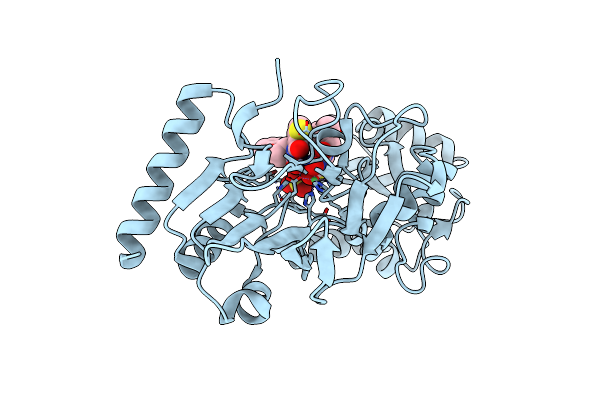 |
Crystal Structure Of Human Dna Cross-Link Repair 1A In Complex With A Cyclic N-Hydroxyurea Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: NI, XOB, R3Z, ZN, SO4 |
 |
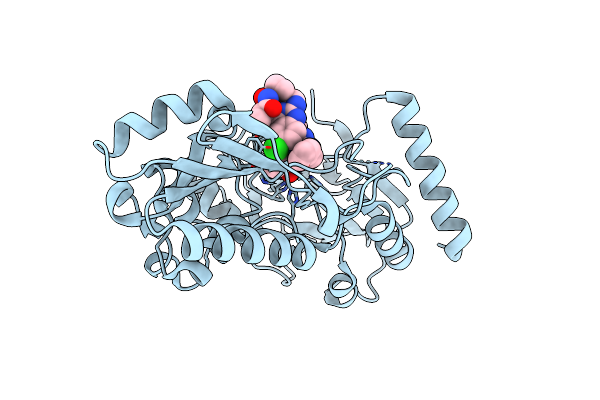
Crystal Structure Of Human Dna Cross-Link Repair 1A In Complex With Hydroxamic Acid Inhibitor (Compound 48).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: U1L, ZN |
 |
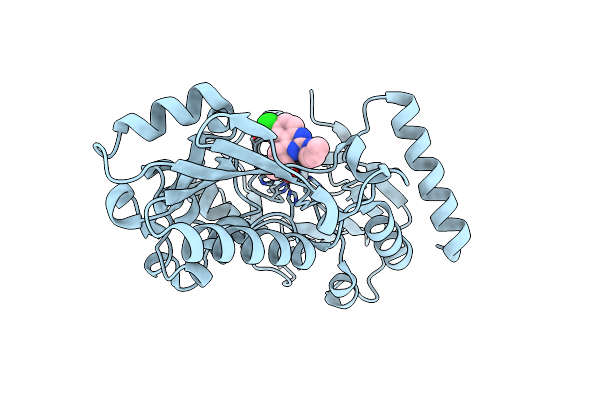
Crystal Structure Of Human Dna Cross-Link Repair 1A In Complex With Hydroxamic Acid Inhibitor (Compound 44).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: U2C, ZN |
 |
Crystal Structure Of Human Dna Cross-Link Repair 1A In Complex With Hydroxamic Acid Inhibitor (Compound 21).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: U2O, ZN |
 |
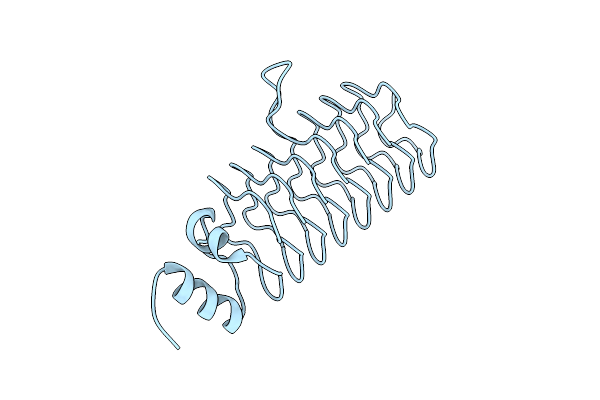
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2016-09-28 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-08-31 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.40 Å Release Date: 2016-08-31 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.27 Å Release Date: 2016-08-31 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-08-31 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2016-08-31 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2016-03-16 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-03-09 Classification: STRUCTURAL PROTEIN Ligands: CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2016-03-09 Classification: STRUCTURAL PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2016-03-09 Classification: STRUCTURAL PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2016-03-09 Classification: STRUCTURAL PROTEIN |

