Search Count: 86
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From The Filarial Nematode W. Bancrofti In Complex With Nadph And [2-({4-[(2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl]Benzene-1-Carbonyl}Amino)-4-Methoxyphenyl]Acetic Acid (Tsd10 Or Oed)
Organism: Wuchereria bancrofti
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: NDP, OED |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From The Filarial Nematode W. Bancrofti In Complex With Nadph And [2-({4-[(2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl]Benzene-1-Carbonyl}Amino)-4-Cyanophenyl]Acetic Acid (Tsd25 Or Ofd)
Organism: Wuchereria bancrofti
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: NAP, OFD |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From The Filarial Nematode W. Bancrofti In Complex With Nadph And Methotrexate (Mtx)
Organism: Wuchereria bancrofti
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: NAP, MTX |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From The Filarial Nematode W. Bancrofti In Complex With Nadph And Antifolate 2-({4-[(2-Amino-4-Oxo-4,7-Dihydro-1H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl]Benzene-1-Carbonyl}Amino)Benzoic Acid (Og7 Or Tsd001)
Organism: Wuchereria bancrofti
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: NAP, OG7 |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 141-156 (Cphalotagdelta)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: NHE |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 154-156 (Cphalotag7_154-156)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: CL |
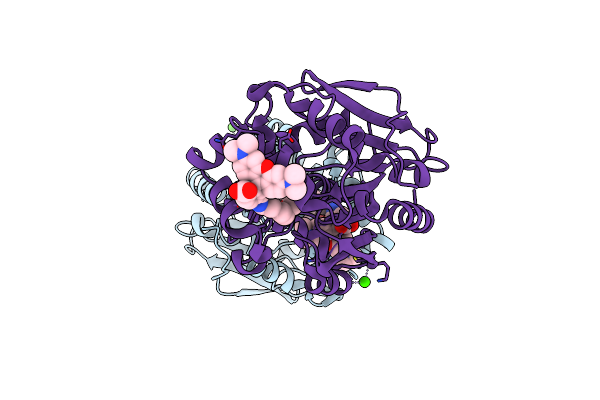 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Bound To A Pentyltrifluoromethanesulfonamide Tetramethylrhodamine Ligand (Tmr-T5)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: IYI, CA |
 |
X-Ray Structure Of The Dead Variant Haloalkane Dehalogenase Halotag7-D106A Bound To A Pentanol Tetramethylrhodamine Ligand (Tmr-Hy5)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: IYL, CL, GOL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Bound To A Pentylmethanesulfonamide Tetramethylrhodamine Ligand (Tmr-S5)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: IYO, GOL |
 |
Organism: Acinetobacter baumannii (strain aye)
Method: ELECTRON MICROSCOPY Release Date: 2021-10-20 Classification: TRANSPORT PROTEIN |
 |
Organism: Acinetobacter baumannii (strain aye)
Method: ELECTRON MICROSCOPY Release Date: 2021-10-20 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-20 Classification: TRANSPORT PROTEIN Ligands: PEG, DXT |
 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-10-20 Classification: TRANSPORT PROTEIN Ligands: FUA |
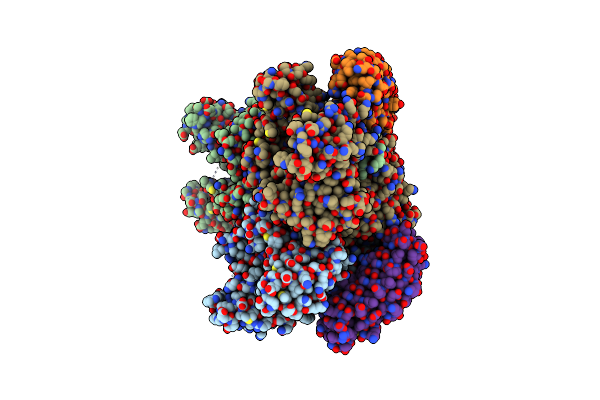 |
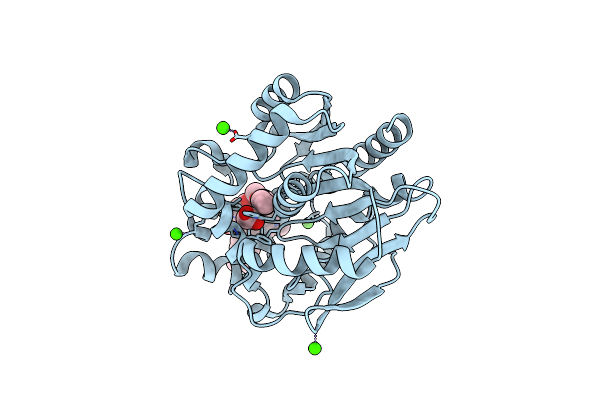
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-10-20 Classification: TRANSPORT PROTEIN Ligands: LFX |
 |
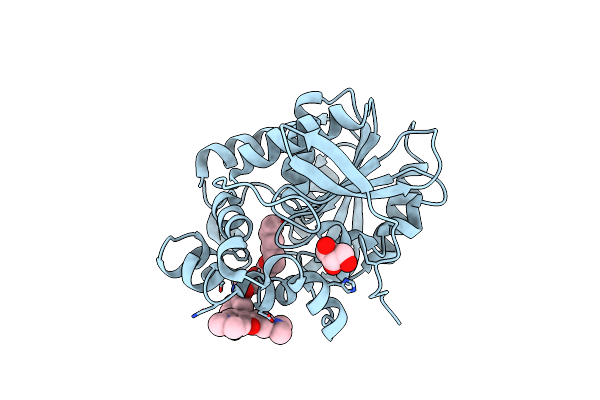
X-Ray Structure Of The Haloalkane Dehalogenase Hob (Halotag7-Based Oligonucleotide Binder) Labeled With A Chloroalkane-Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-04-21 Classification: HYDROLASE Ligands: OEH, ACT, CA |
 |
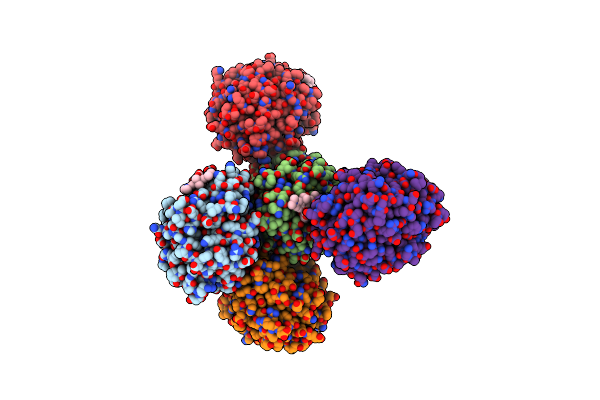
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Labeled With A Chloroalkane-Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: OEH, CL, GOL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Labeled With A Chloroalkane-Carbopyronine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: OEK, CL |
 |
Crystal Structure Of Snap-Tag Labeled With A Benzyl-Tetramethylrhodamine Fluorophore
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-03-31 Classification: TRANSFERASE Ligands: ZN, OGQ, EDO |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-12-18 Classification: METAL BINDING PROTEIN Ligands: GOL, PO4, K |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-12-18 Classification: METAL BINDING PROTEIN Ligands: FEC, GOL |

