Search Count: 16
 |
Organism: Methylophaga sulfidovorans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-07-31 Classification: TRANSFERASE |
 |
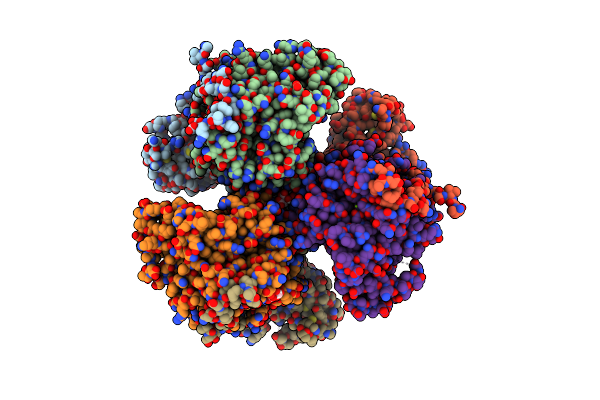
Organism: Ananas comosus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSFERASE |
 |
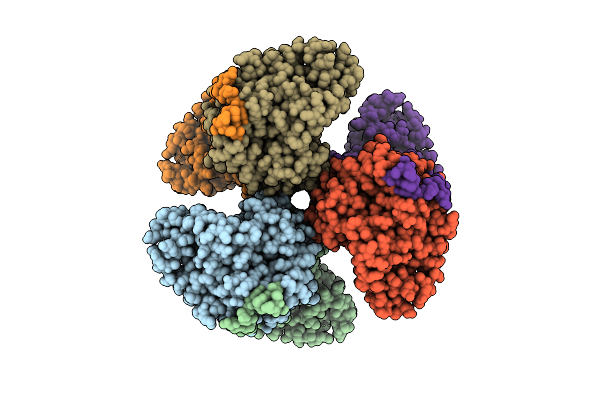
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2024-04-24 Classification: TRANSFERASE Ligands: SO4, CIT, MG |
 |
Structure Of Hexameric Subcomplexes (Truncation Delta2-6) Of The Fractal Citrate Synthase From Synechococcus Elongatus Pcc7942
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: TRANSFERASE |
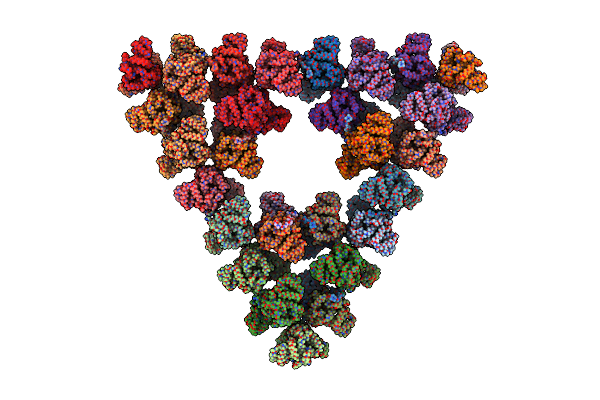 |
Pseudoatomic Model Of A Second-Order Sierpinski Triangle Formed By The Citrate Synthase From Synechococcus Elongatus
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: TRANSFERASE |
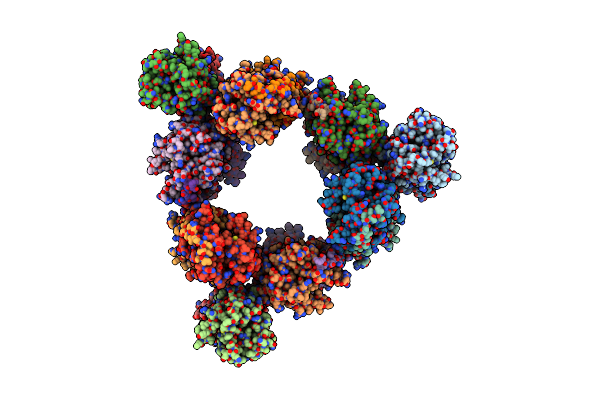 |
Structure Of A First Order Sierpinski Triangle Formed By The H369R Mutant Of The Citrate Synthase From Synechococcus Elongatus
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: TRANSFERASE |
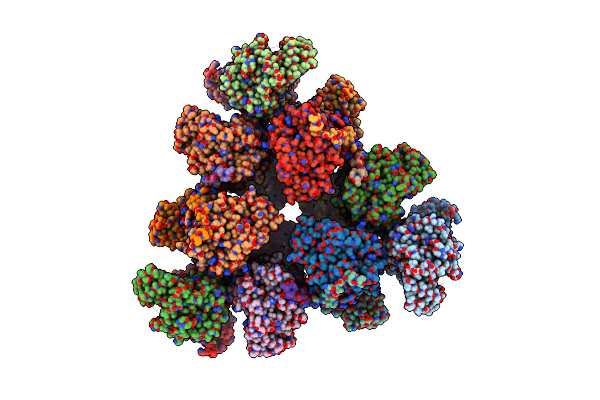 |
Structure Of A First Level Sierpinski Triangle Formed By A Citrate Synthase
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: TRANSFERASE |
 |
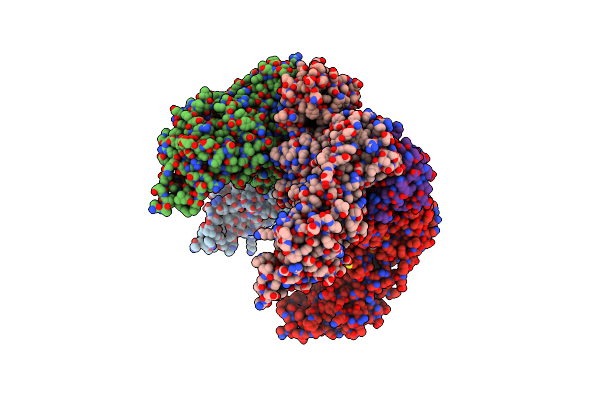
L8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction Of The Last Common Ancestor Of Form I'' And Form I Rubiscos
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
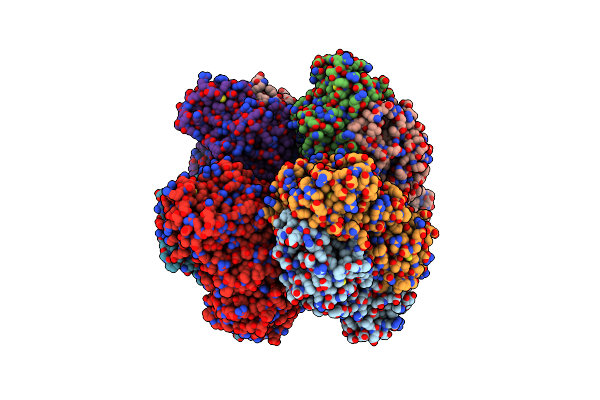
L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction Of The Last Common Ancestor Of Ssu-Bearing Form I Rubiscos
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
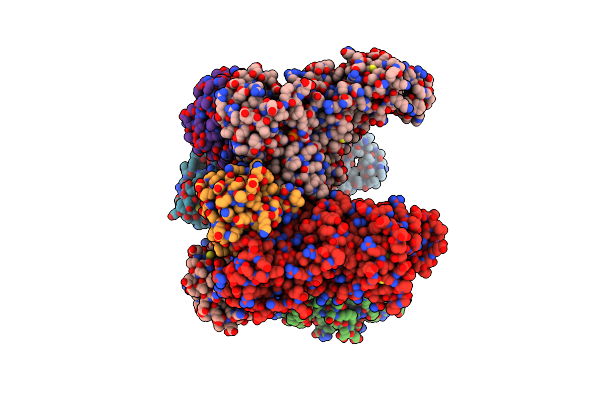
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8 Complex
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8S8 Complex
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8 Complex With Substitution E170N
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8S8 Complex With Substitution E170N
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Fiber-Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Organism: Aromatoleum aromaticum (strain ebn1), Aromatoleum aromaticum ebn1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-09-26 Classification: RNA BINDING PROTEIN Ligands: GOL, MN, TLA |
 |
Organism: Aromatoleum aromaticum (strain ebn1), Aromatoleum aromaticum ebn1
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2018-09-26 Classification: RNA BINDING PROTEIN Ligands: TLA, GOL, MN |

