Search Count: 45
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Imp And The Inhibitor P221
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, GOL, ZO7, K, EDO |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Imp And The Inhibitor P176
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: IMP, ZO4, K |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Campylobacter Jejuni In The Complex With Inhibitor Oxanosine Monophosphate
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2018-10-24 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: JQS, SO4, CL, MPD, K |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Inhibitor Oxanosine Monophosphate
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-10-24 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: JQS, K, EDO, PGE, PEG |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P225
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2017-05-24 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8KY |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P221
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8N1, MPD, ACY, FMT, MRD |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P182
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8L1, K, GOL |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P200
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8L4, PGE, PEG, EDO, MG, K, PG4 |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Imp And The Inhibitor P178
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE Ligands: IMP, 8LA, K, EDO, PO4, GOL, PEG |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Imp And The Inhibitor P200
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8L4, K |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P178
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8LA, K, MPD, FMT, SO4 |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Campylobacter Jejuni In The Complex With Imp And The Inhibitor P225
Organism: Campylobacter jejuni subsp. jejuni, Campylobacter jejuni subsp. jejuni cg8486
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2017-03-01 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8KY, K, SO4, EDO, GOL, CL |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Campylobacter Jejuni In The Complex With Inhibitor P200
Organism: Campylobacter jejuni subsp. jejuni, Campylobacter jejuni subsp. jejuni cg8486
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2017-03-01 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8L4, EDO, K |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Campylobacter Jejuni In The Complex With Inhibitor P176
Organism: Campylobacter jejuni subsp. jejuni, Campylobacter jejuni subsp. jejuni cg8486
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-03-01 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8L7, K |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With A Product Imp And The Inhibitor P182
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-03-01 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8L1, K, GOL |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P176
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-01 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: 8L7, IMP, MPD, ACY, MRD |
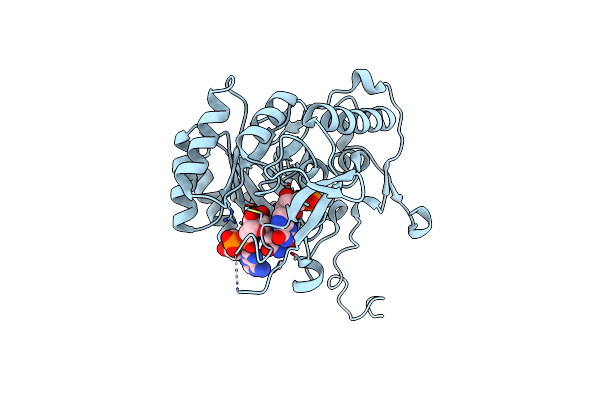 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Mycobacterium Tuberculosis In The Complex With Xmp And Nad
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-06-17 Classification: OXIDOREDUCTASE Ligands: XMP, NAD |
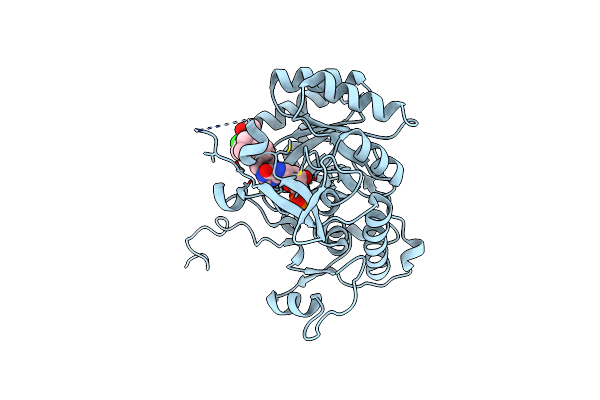 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Mycobacterium Tuberculosis In The Complex With Imp And The Inhibitor P41
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-06-17 Classification: Oxidoreductase/Oxidoreductase inhibitor Ligands: IMP, 4QO |
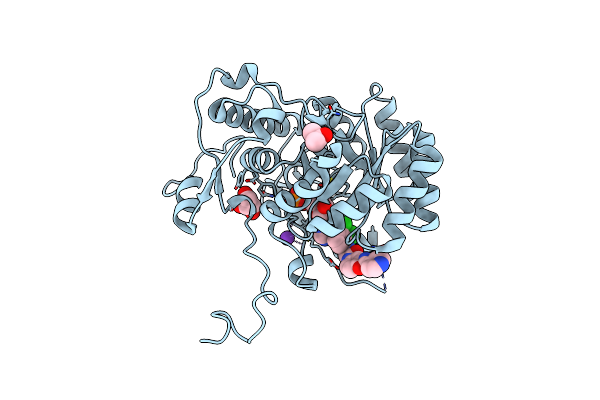 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Mycobacterium Tuberculosis In The Complex With Imp And The Inhibitor Q67
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-06-17 Classification: Oxidoreductase/Oxidoreductase inhibitor Ligands: IMP, Q67, K, GOL, PGO |
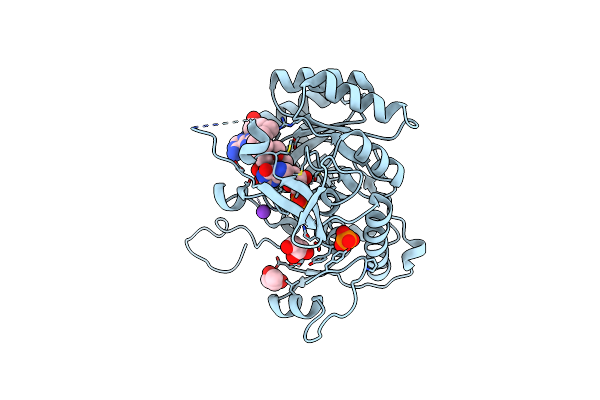 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Mycobacterium Tuberculosis In The Complex With Imp And The Inhibitor Mad1
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-06-17 Classification: Oxidoreductase/Oxidoreductase inhibitor Ligands: IMP, KP3, K, PO4, GOL, PGO |

