Search Count: 17
 |
Organism: Turnip yellows virus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN |
 |
Organism: Potato leafroll virus
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Potato leafroll virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-01-19 Classification: VIRAL PROTEIN Ligands: BR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2018-01-10 Classification: TRANSPORT PROTEIN |
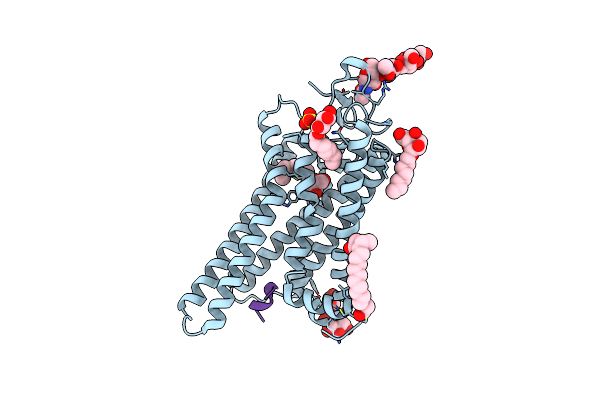 |
Crystal Structure Of The Active G-Protein-Coupled Receptor Opsin In Complex With The Finger-Loop Peptide Derived From The Full-Length Arrestin-1
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2014-09-17 Classification: SIGNALING PROTEIN Ligands: BOG, PLM, SO4, ACT |
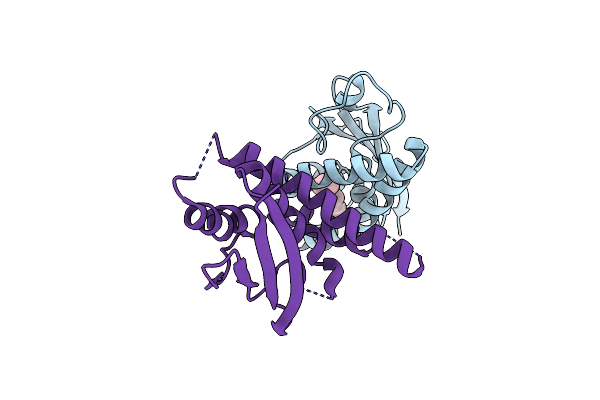 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-04-21 Classification: TRANSPORT PROTEIN Ligands: PLM |
 |
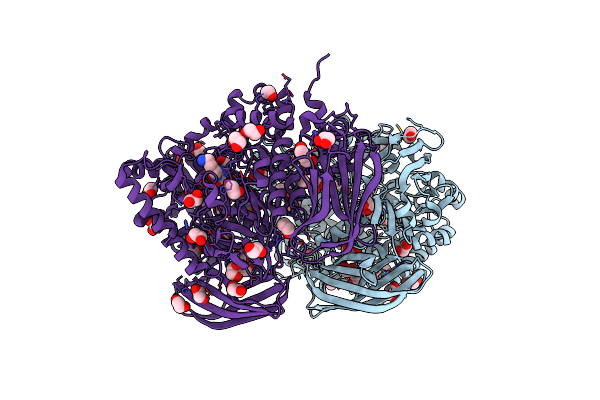
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-08 Classification: HYDROLASE Ligands: 15A, EDO, BR, CL |
 |
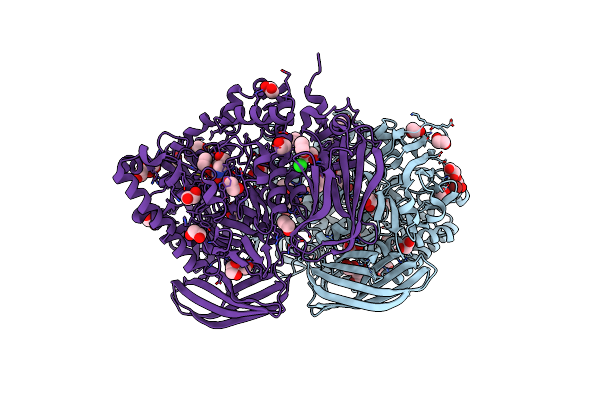
Transition-State Mimicry In Mannoside Hydrolysis: Characterisation Of Twenty Six Inhibitors And Insight Into Binding From Linear Free Energy Relationships And 3-D Structure
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-04-08 Classification: HYDROLASE Ligands: 17B, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: IFL, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: MNM, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: MVL, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: VBZ, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: NHV, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: EDO, NOY, BR, CL |
 |
Crystal Structure Of Oxyb, A Cytochrome P450 Implicated In An Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis
Organism: Amycolatopsis orientalis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2002-12-11 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Oxyb, A Cytochrome P450 Implicated In An Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis
Organism: Amycolatopsis orientalis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-12-11 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Oxyb, A Cytochrome P450 Implicated In An Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis
Organism: Amycolatopsis orientalis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-12-11 Classification: OXIDOREDUCTASE Ligands: HEM |

