Search Count: 51
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of Surfactant Protein-A Dedn Mutant (E171D/P175E/R197N/K203D)
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: HEZ, CA, CL |
 |
Crystal Structure Of Surfactant Protein-A Dedn Mutant (E171D/P175E/R197N/K203D) Complexed With Inositol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: CA, INS, HEZ, CL |
 |
Crystal Structure Of Surfactant Protein-A Dedn Mutant (E171D/P175E/R197N/K203D) Complexed With Mannose
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: CA, MAN, HEZ, CL |
 |
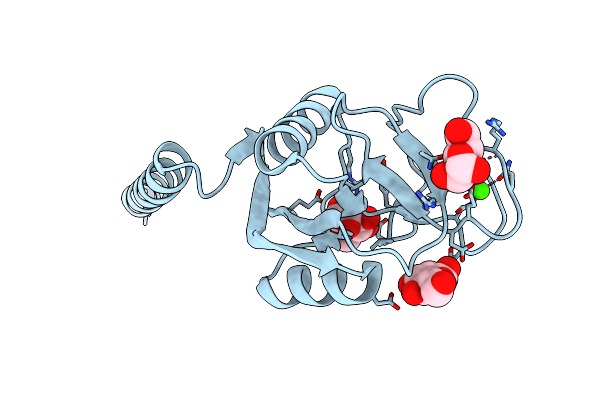
Crystal Structure Of Surfactant Protein-A Ded Mutant (E171D/P175E/K203D) Complexed With Inositol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: INS, CA |
 |
Crystal Structure Of Surfactant Protein-A Ded Mutant (E171D/P175E/K203D) Complexed With Mannose
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: MAN, CA |
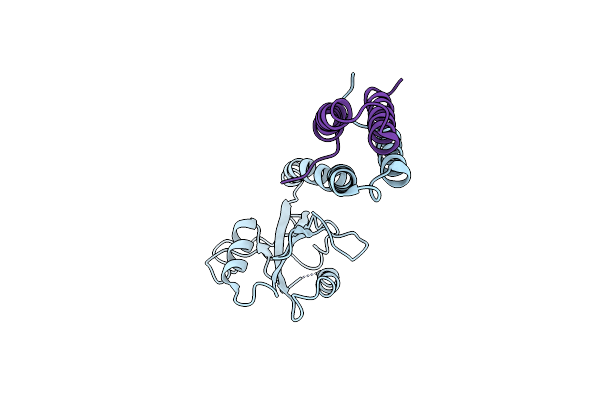 |
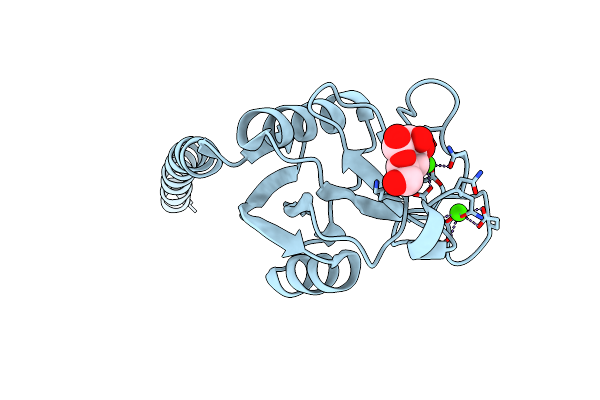
Organism: Legionella pneumophila subsp. pneumophila, Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-05-22 Classification: PROTEIN BINDING |
 |
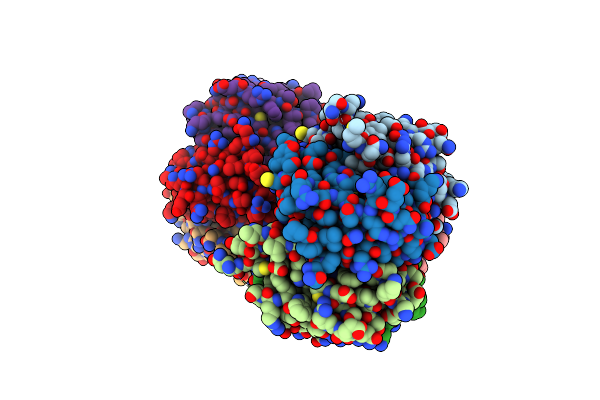
Structure Of Porcine Surfactant Protein D Neck And Carbohydrate Recognition Domain Complexed With Mannose
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-06-20 Classification: SUGAR BINDING PROTEIN Ligands: CA, BMA |
 |
Human Nucleoplasmin (Npm2): A Histone Chaperone In Oocytes And Early Embryos
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-09-21 Classification: CHAPERONE |
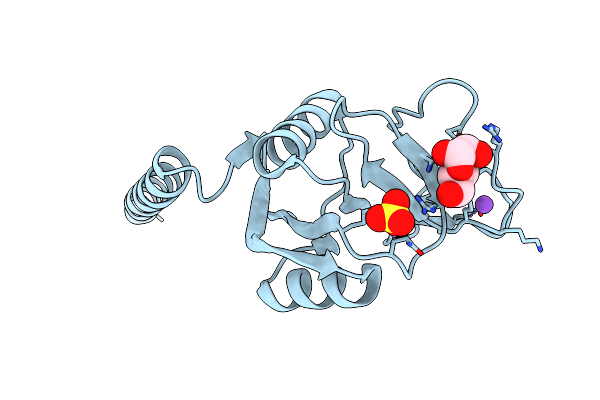 |
Crystal Structure Of Rat Surfactant Protein A Neck And Carbohydrate Recognition Domain (Ncrd) Complexed With Mannose
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-11-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, NA, SO4, MAN |
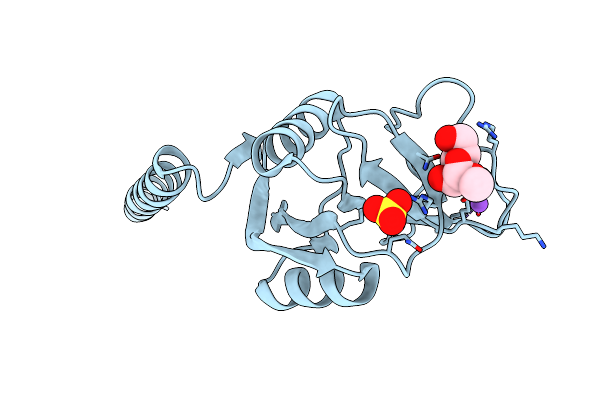 |
Surfactant Protein A Neck And Carbohydrate Recognition Domain (Ncrd) Complexed With Alpha-Methylmannose
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-11-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, NA, SO4, MMA |
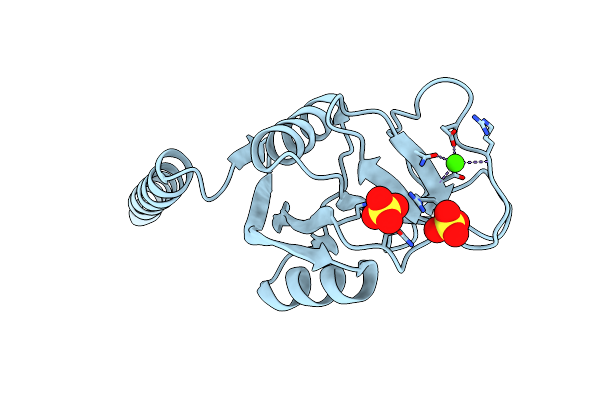 |
Surfactant Protein-A Neck And Carbohydrate Recognition Domain (Ncrd) In The Absence Of Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-11-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, SO4 |
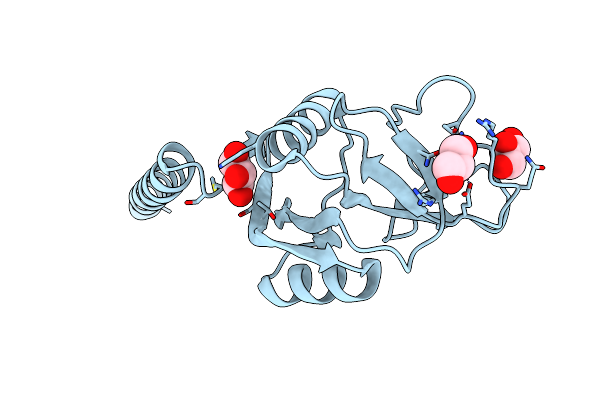 |
Surfactant Protein-A Neck And Carbohydrate Recognition Domain (Ncrd) Complexed With Glycerol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-11-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL |
 |
Organism: Legionella pneumophila, Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-04-28 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Interacting Domains Of Icmr And Icmq (Seleno-Derivative)
Organism: Legionella pneumophila, Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-04-28 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of The Y247S/Y251S Mutant Of Phosphatidylinositol-Specific Phospholipase C From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-04-14 Classification: LYASE Ligands: ZN |
 |
Crystal Structure Of The Myo-Inositol Bound Y247S/Y251S Mutant Of Phosphatidylinositol-Specific Phospholipase C From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-04-14 Classification: LYASE Ligands: INS, ZN |
 |
Crystal Structure Of The Y246S/Y247S/Y248S/Y251S Mutant Of Phosphatidylinositol-Specific Phospholipase C From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2009-04-14 Classification: LYASE Ligands: MN |
 |
Crystal Structure Of The C2 Domain Of Bovine Lactadherin At 1.67 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2007-12-25 Classification: BLOOD CLOTTING, CELL ADHESION |
 |
Structure Of The W47A/W242A Mutant Of Bacterial Phosphatidylinositol-Specific Phospholipase C
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2007-02-13 Classification: LYASE |

