Search Count: 786
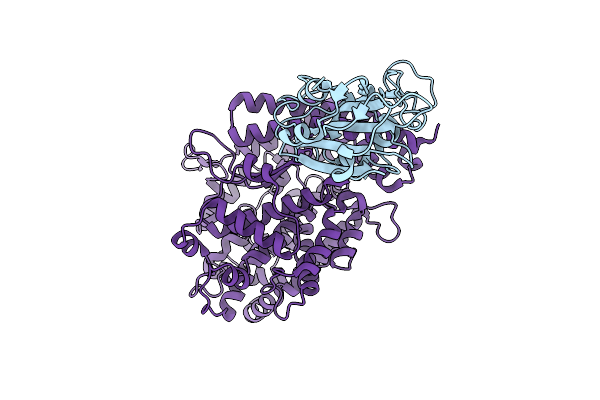 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
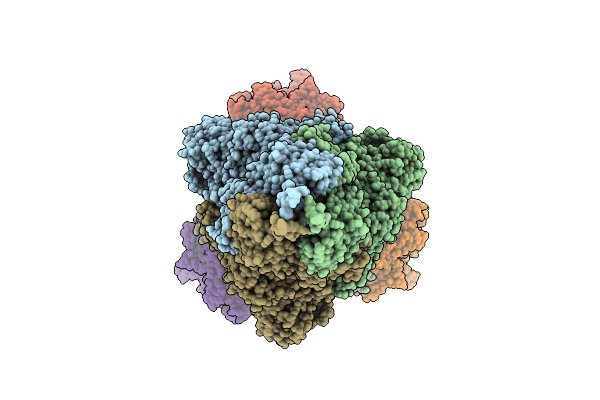 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
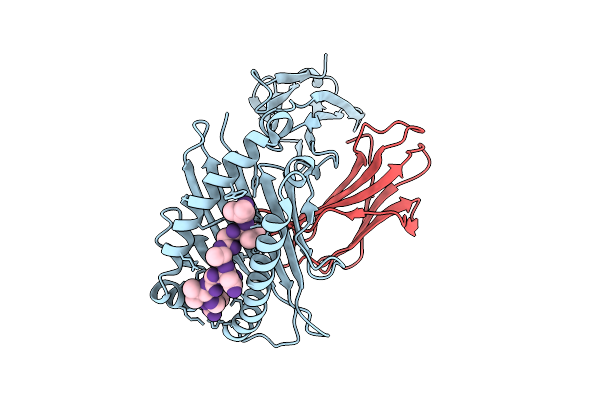 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12D 9-Mer Peptide (Vvgadgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
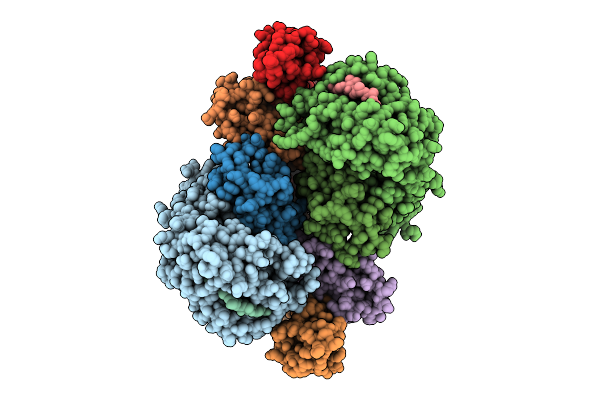 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12A 10-Mer Peptide (Vvvgaagvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12S 10-Mer Peptide (Vvvgasgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
Solution Structure Of The Human Prostate Stem Cell Antigen (Psca), Water-Soluble Domain
|
 |
Crystal Structure Of Human Shiftless (Sfl) Containing Phosphorylation Sites Ser249, Thr250, Thr253 And Ser256
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-19 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Nocardioides
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Mogibacterium pumilum
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2025-11-05 Classification: RNA |
 |
Crystal Structure Of Arabidopsis Thaliana Ing1 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
 |
Crystal Structure Of Arabidopsis Thaliana Ing2 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.97 Å Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: HVG, A1ENS |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.14 Å Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: GLU |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: Z99 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: A1ENR, HVG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Transaldolase Aprg From Streptoalloteichus Tenebrarius
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: PEG |
 |
Complex Crystal Structure Of Transaldolase Aprg With Glcnac From Streptoalloteichus Tenebrarius
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: A1EJ3, A1EJ2, PEG, NAG, A1EJ0 |
 |
Complex Crystal Structure Of Transaldolase Aprg With Galnac From Streptoalloteichus Tenebrarius
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: PEG |

