Search Count: 866
 |
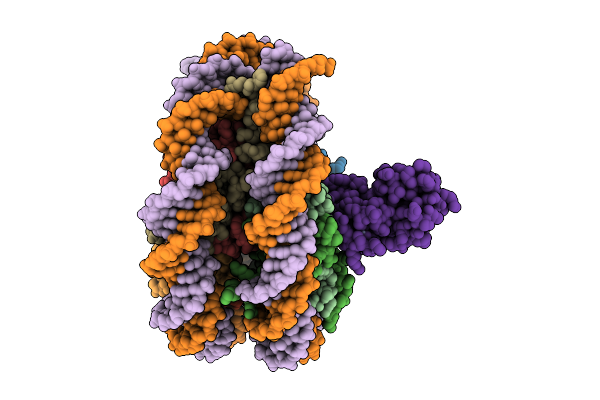
Organism: Leptospirillum ferriphilum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of The Type Ii Secretion System Protein From Acidithiobacillus Caldus
Organism: Acidithiobacillus caldus (strain sm-1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TOXIN |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
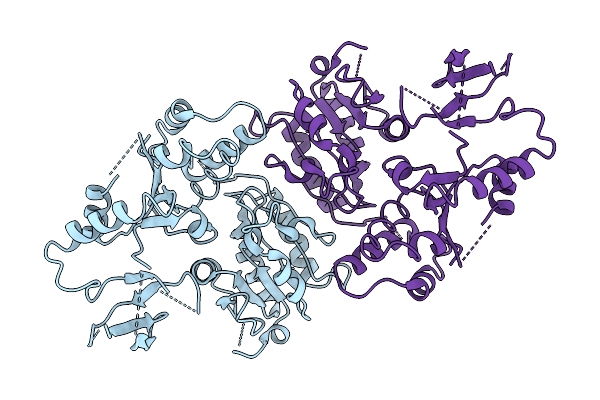
Dihydrolipoamide Acetyltransferase (E2) Psbd In Complex With The Pyruvate Dehydrogenase (E1) Binding Domain From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PROTEIN BINDING |
 |
Dihydrolipoyl Dehydrogenase (E3) In Complex With The Binding Domain Of Dihydrolipoamide Acetyltransferase (E2) From The E. Coli Pyruvate Dehydrogenase Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: FLAVOPROTEIN Ligands: FAD |
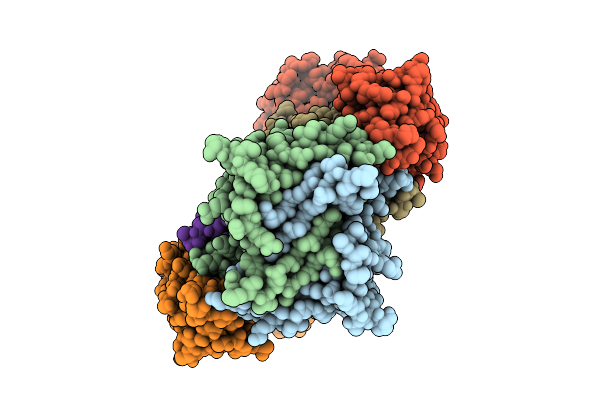 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: VIRAL PROTEIN |
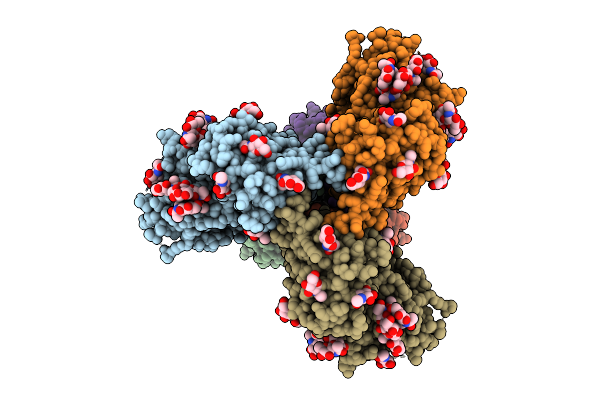 |
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN Ligands: NAG |
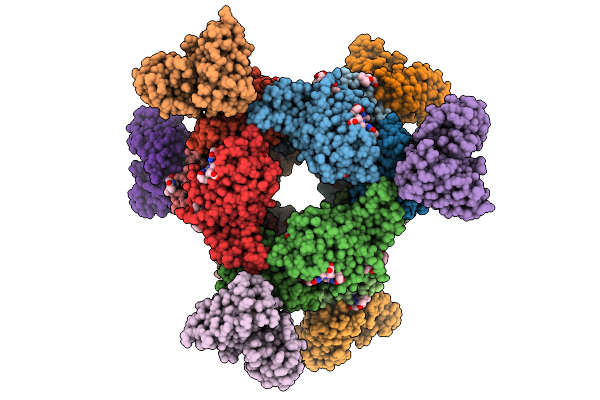 |
Organism: Mus musculus, Neopyropia tenera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: PEB, PUB |
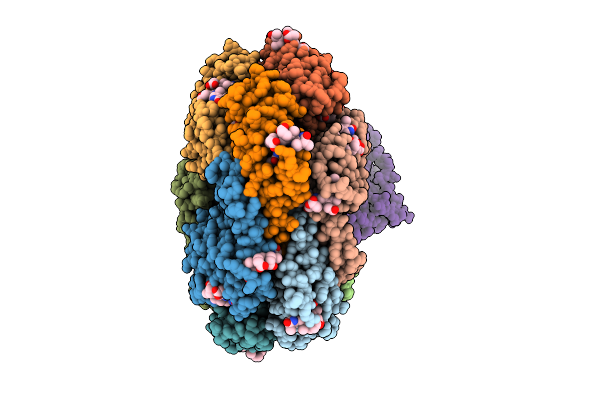 |
Organism: Mus musculus, Ceramium secundatum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: PEB, PUB |
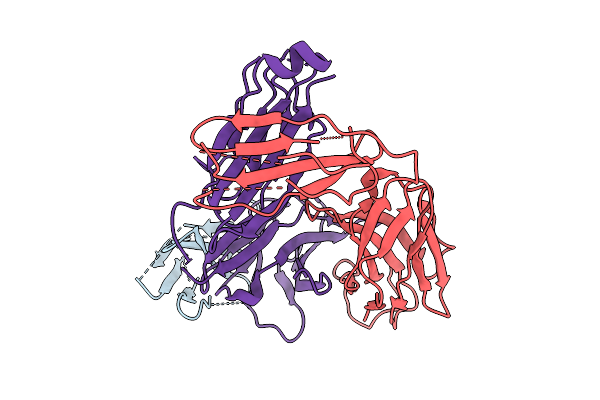 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM |
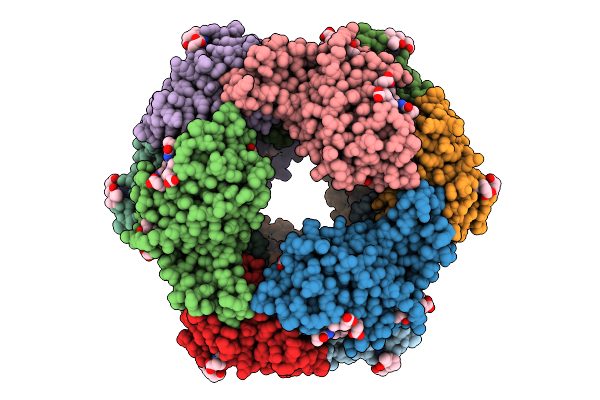 |
Organism: Pyropia tenera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: PEB, PUB |
 |
Organism: Homo sapiens, Pyropia tenera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: PEB, PUB |
 |
Organism: Human metapneumovirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: A1IUN, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: A1IUM, EDO |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: DNA BINDING PROTEIN |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: CL |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: CL |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: CL, NA |

