Search Count: 42
 |
Organism: Thiopseudomonas alkaliphila, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: DNA BINDING PROTEIN/DNA Ligands: PO4 |
 |
Crystal Structure Of Human Transthyretin In Complex With 3-O-Methyltolcapone Analogue 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-01-17 Classification: LIPID BINDING PROTEIN Ligands: U1F |
 |
Crystal Structure Of Human Transthyretin In Complex With 3-O-Methyltolcapone Analogue 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2024-01-17 Classification: LIPID BINDING PROTEIN Ligands: TQ0 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2023-09-20 Classification: TRANSLATION Ligands: MG, GDP, Y7C |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-08-31 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, IAZ, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-08-31 Classification: CELL CYCLE Ligands: GTP, MG, GDP, MES, NW6, ACP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-04-20 Classification: LYASE Ligands: ZN, 8EK, K |
 |
U34-Trna Thiolase Ncsa From Methanococcus Maripaludis With Its [4Fe-4S] Cluster
Organism: Methanococcus maripaludis (strain s2 / ll)
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2020-08-26 Classification: RNA BINDING PROTEIN Ligands: ZN, SF4, SO4 |
 |
 |
Ecv-1 From Echinicola Vietnamensis. Environmental Metallo-Beta-Lactamases Exhibit High Enzymatic Activity Under Zinc Deprivation
Organism: Echinicola vietnamensis
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2020-05-27 Classification: ANTIMICROBIAL PROTEIN Ligands: EDO, ZN |
 |
Myo-1 From Myroides Odoratimimus. Environmental Metallo-Beta-Lactamases Exhibit High Enzymatic Activity Under Zinc Deprivation
Organism: Myroides odoratimimus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2020-05-27 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN, MG |
 |
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2019-05-22 Classification: ISOMERASE Ligands: SO4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Propylamycin And Bound To Mrna And A-, P-, And E-Site Trnas At 2.75A Resolution
Organism: Escherichia coli, Enterobacteria phage t4, Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-04-17 Classification: RIBOSOME/ANTIBIOTIC Ligands: MG, LUJ, K, ZN, SF4 |
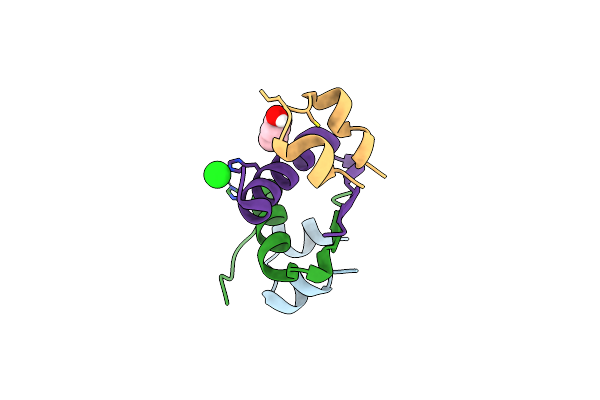 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-06-13 Classification: HORMONE Ligands: ZN, IPH, CL |
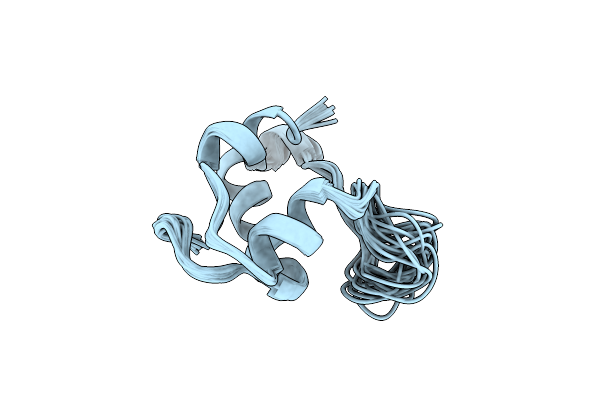 |
Solution Structure And Dynamics Of An Ultra-Stable Single-Chain Insulin Analog Studies Of An Engineered Monomer And Implications For Receptor Binding
|
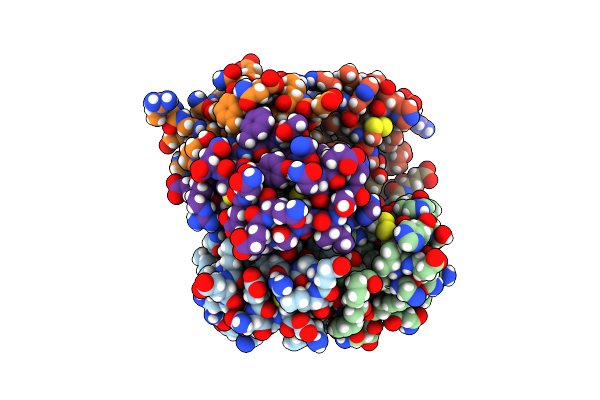 |
An Ultra-Stable Single-Chain Insulin Analog Resists Thermal Inactivation And Exhibits Biological Signaling Duration Equivalent To The Native Protein
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-11-15 Classification: HORMONE |
 |
Organism: Pelophylax esculentus
Method: SOLUTION NMR Release Date: 2017-10-04 Classification: ANTIMICROBIAL PROTEIN |
 |
Nmr Solution Structure Of The Nav1.7 Selective Spider Venom-Derived Peptide Pn3A
|
 |
Structure Of Human Sirt2 In Complex With 1,2,4-Oxadiazole Inhibitor And Adp Ribose.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: ZN, AR6, 7KE, EDO, DMS, GOL, ACT |
 |
Crystal Structure Of The N-Terminal N-Formyltransferase Domain (Residues 1-306) Of Escherichia Coli Arna In Complex With Udp-Ara4N And Folinic Acid
Organism: Escherichia coli h736
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-05-25 Classification: TRANSFERASE Ligands: G3N, FNX |

