Search Count: 13
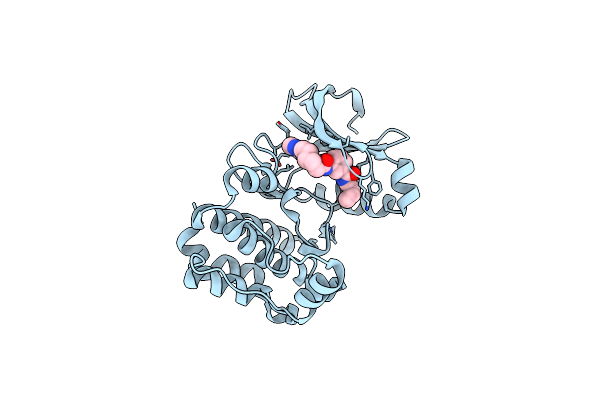 |
Crystal Structure Of Aurora A Complexed With An Inhibitor Discovered Through Site-Directed Dynamic Tethering
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-07-08 Classification: TRANSFERASE Ligands: FXG |
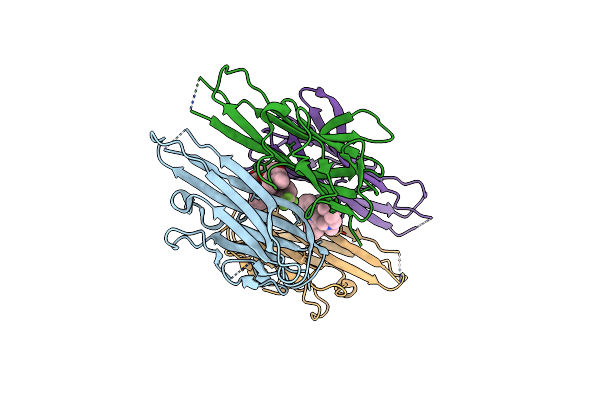 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-11-29 Classification: CYTOKINE Ligands: 307 |
 |
Crystal Structure Of Phage T4 Lysozyme Mutant Y24A/Y25A/T26A/I27A/C54T/C97A
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2004-10-19 Classification: HYDROLASE Ligands: BME |
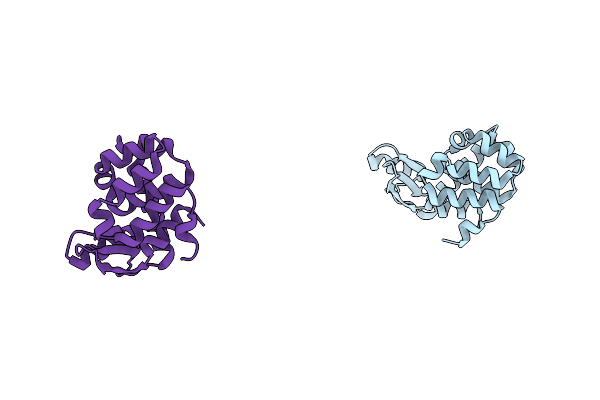 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-10-19 Classification: HYDROLASE |
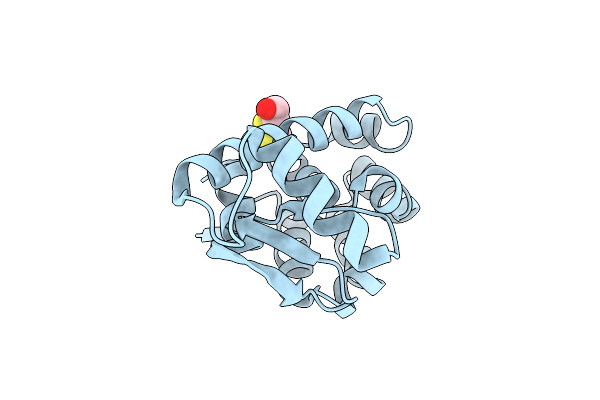 |
Crystal Structure Of Phage T4 Lysozyme Mutant R14A/K16A/I17A/K19A/T21A/E22A/C54T/C97A
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2004-10-19 Classification: HYDROLASE Ligands: CL, BME |
 |
Crystal Structure Of Phage T4 Lysozyme Mutant L32A/L33A/T34A/C54T/C97A/E108V
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-10-19 Classification: HYDROLASE Ligands: PO4, CL, BME |
 |
Ptp1B R47C Modified At C47 With N-[4-(2-{2-[3-(2-Bromo-Acetylamino)-Propionylamino]-3-Hydroxy-Propionylamino}-Ethyl)-Phenyl]-Oxalamic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2003-05-06 Classification: HYDROLASE Ligands: FG1 |
 |
Crystal Structure Of The Ptp1B Complexed With Sp7343-Sp7964, A Ptyr Mimetic
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-04-01 Classification: HYDROLASE Ligands: MG, 964 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2000-09-20 Classification: LYASE Ligands: NI |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2000-09-20 Classification: LYASE Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-09-20 Classification: LYASE Ligands: CO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-09-20 Classification: LYASE Ligands: CD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2000-09-20 Classification: LYASE |

