Search Count: 235
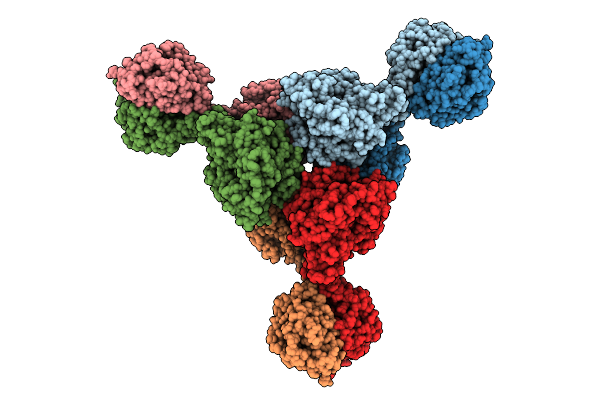 |
Crispr-Associated Deaminase Cad1 In Ca4 Bound In Hexamer Form Refined Against The Consensus Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: ANTIVIRAL PROTEIN/RNA Ligands: ZN |
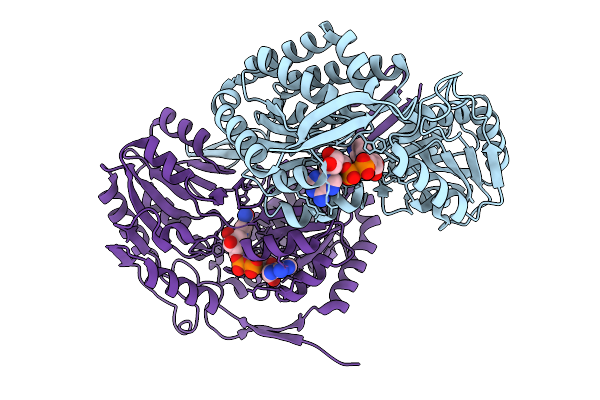 |
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Oryza sativa subsp. japonica
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSPORT PROTEIN |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSPORT PROTEIN Ligands: PO4 |
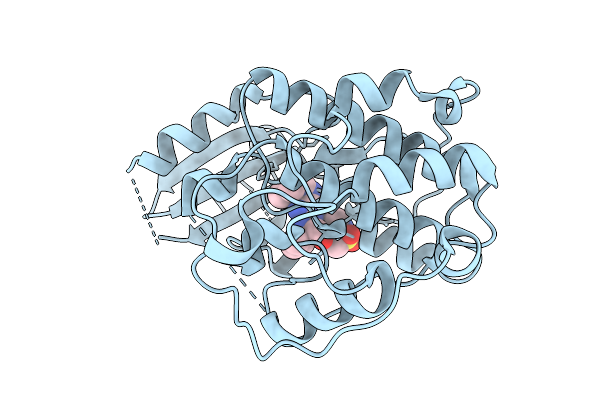 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE Ligands: A1INI |
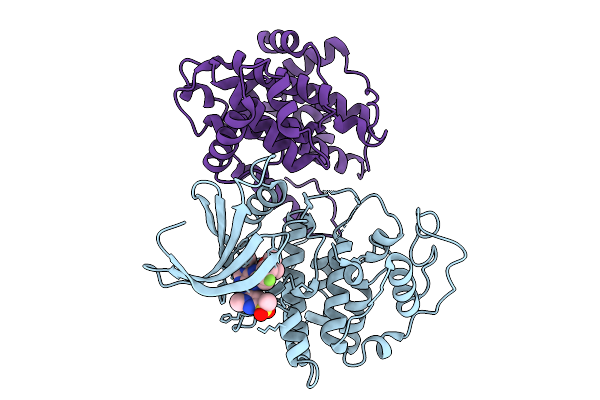 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE Ligands: A1IOI |
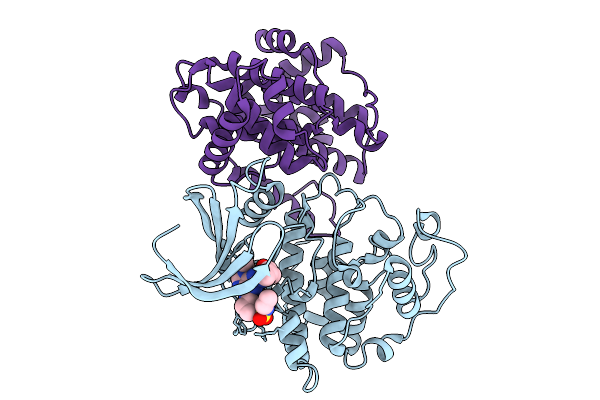 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE Ligands: A1IOP |
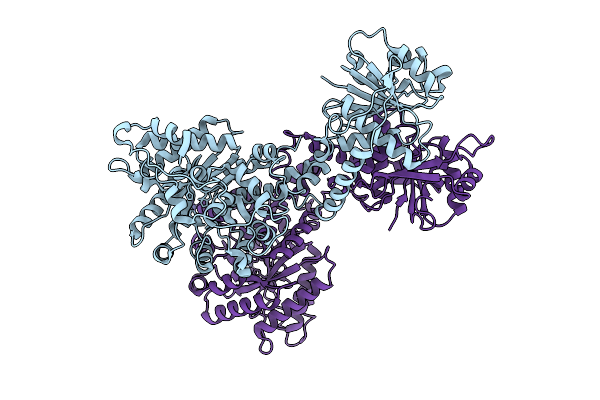 |
Cryoem Structure Of Cad1 In Apo Form, Symmetry Expanded Dimer, Refined Against A Composite Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ZN |
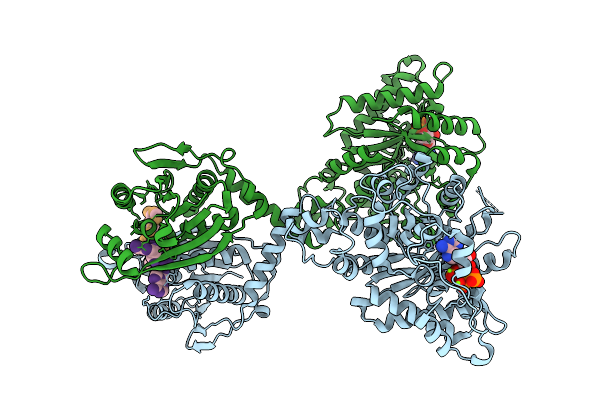 |
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Symmetry Expanded Dimer Refined Against A Composite Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
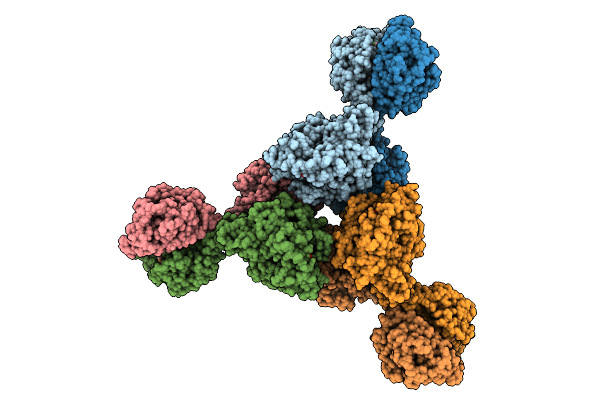 |
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Hexamer With Three Intact Dimers
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
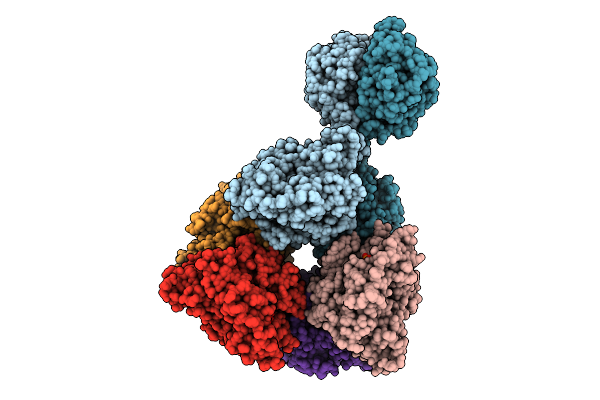 |
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Hexamer With One Intact Dimer
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
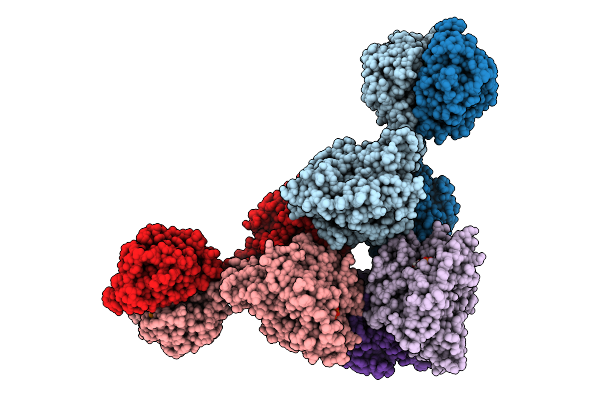 |
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Hexamer With Two Intact Dimers
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
 |
Organism: Pseudomonas phage vb_pa32_gums
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: UNKNOWN FUNCTION |
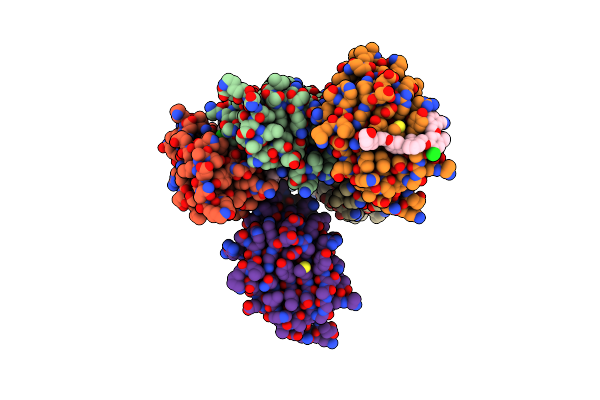 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: IMMUNE SYSTEM Ligands: A1EJ6 |
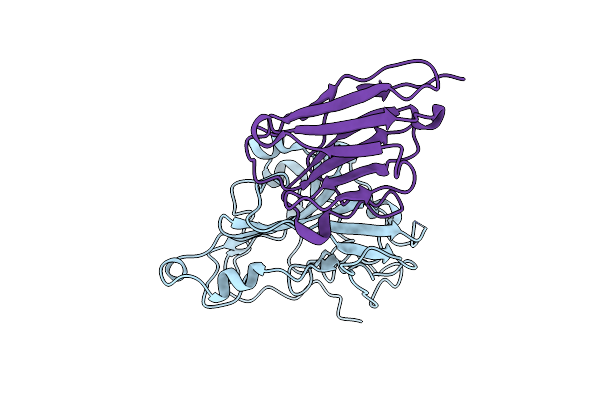 |
Organism: Human alphaherpesvirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:3.72 Å Release Date: 2025-05-28 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Human alphaherpesvirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-05-28 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: MEMBRANE PROTEIN Ligands: A1LXR |
 |
Organism: Homo sapiens, Mus musculus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: 16C, CLR |
 |
Beta-Keto Acid Cleavage Enzyme From Paracoccus Denitrificans With Bound Acetoacetate And Acetyl-Coa
Organism: Paracoccus denitrificans pd1222
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-16 Classification: LYASE Ligands: ACO, AAE, ZN |
 |
Organism: Chloroflexus aurantiacus j-10-fl
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: HEC, SF4, F3S |

