Search Count: 363
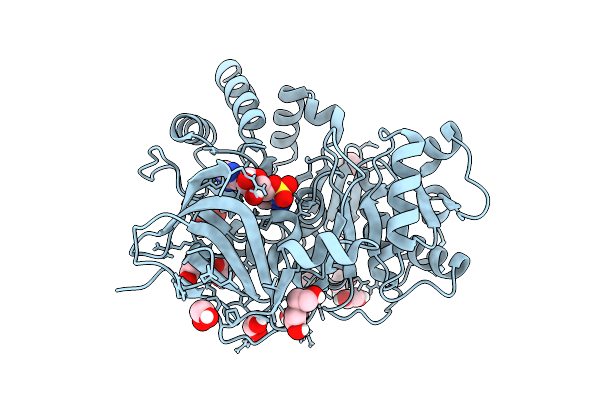 |
High-Resolution Crystal Structure Of Methyl-2,3-Diamino Propanoic Acid-Ams Inhibitor Bound Adenylation Domain (A3) From Sulfazecin Nonribosomal Peptide Synthetase Sulm
Organism: Paraburkholderia acidicola
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: A1BVA, EDO, PEG |
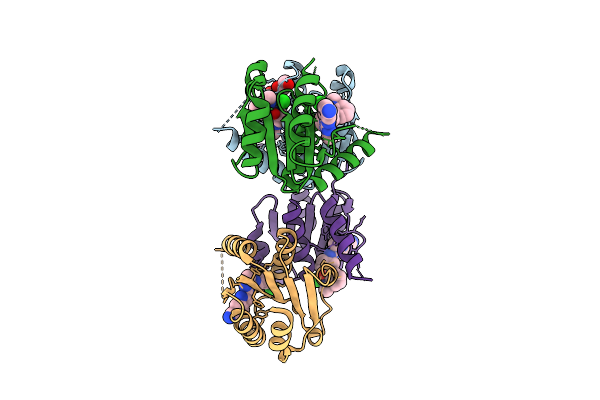 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: EOH, EDO, A1I1M, CL |
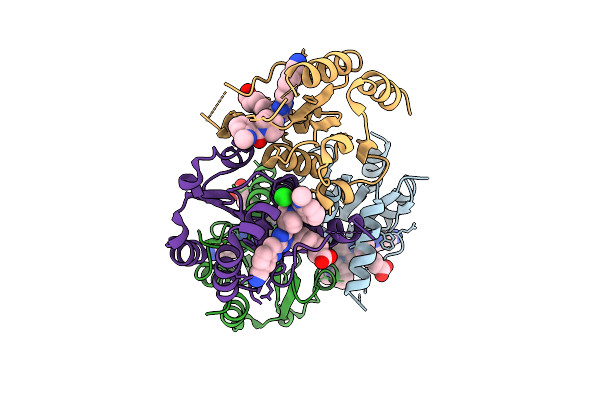 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: CL, A1I1U |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1JFX |
 |
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 2 (H2-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, GDP, PO4 |
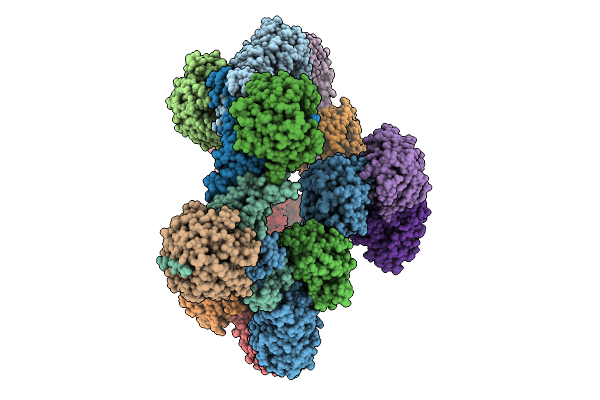 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 1 (H1-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, PO4, GDP |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: PROTEIN FIBRIL Ligands: BDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSFERASE Ligands: SAM, A1CBE, GOL |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
High-Resolution Crystal Structure Of 2,3-Diamino Propanoic Acid Bound Adenylation Domain (A3) From Sulfazecin Nonribosomal Peptide Synthetase Sulm
Organism: Paraburkholderia acidicola
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-05-28 Classification: LIGASE Ligands: EDO, DPP |
 |
Crystal Structure Of Human Soluble Epoxide Hydrolase C-Terminal Domain In Complex With A Benzohomoadamantane-Based Urea Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: A1H8Y |
 |
Structure Of Human Rnf213 Bound To The Secreted Effector Ipah1.4 From Shigella Flexneri
Organism: Homo sapiens, Shigella flexneri 5a str. m90t
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: ANTIMICROBIAL PROTEIN Ligands: ATP, MG, ZN |
 |
Structure Of Human Rnf213 Bound To The Secreted Effector Ipah2.5 From Shigella Flexneri
Organism: Shigella flexneri 5a str. m90t, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: ANTIMICROBIAL PROTEIN Ligands: ATP, MG, ZN |

