Search Count: 36
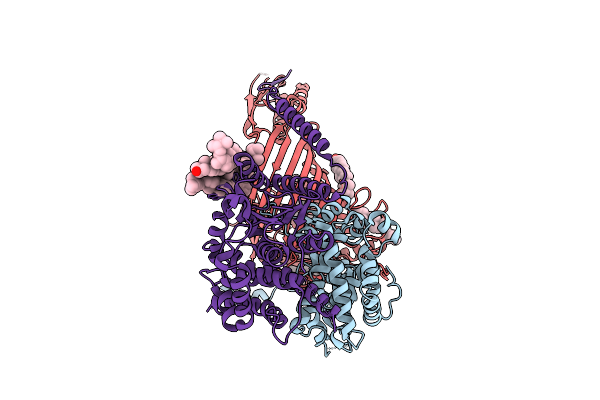 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
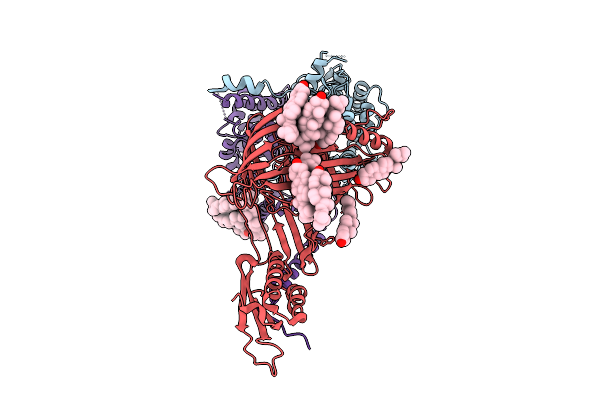 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
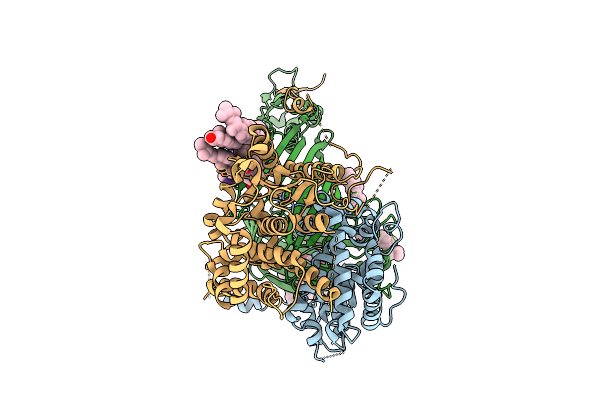 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg3350)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: 12P |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg2000Mme)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: 12P |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg550Dme)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: HZA |
 |
Crystal Structure Of Anti-Peg Antibody M11 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg3350)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: SO4, PE8 |
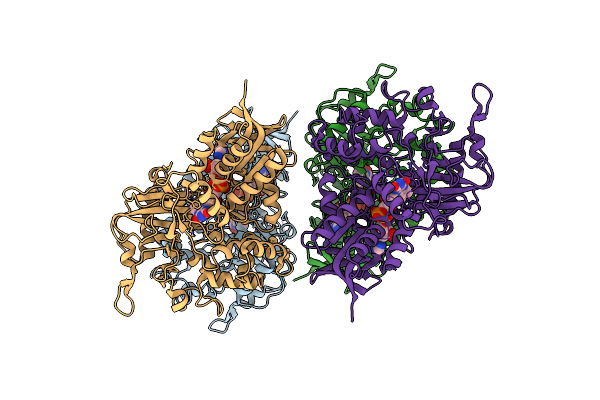 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-10-04 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.52 Å Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: CL, CLR, LBN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.63 Å Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: SO4, CLR, LBN |
 |
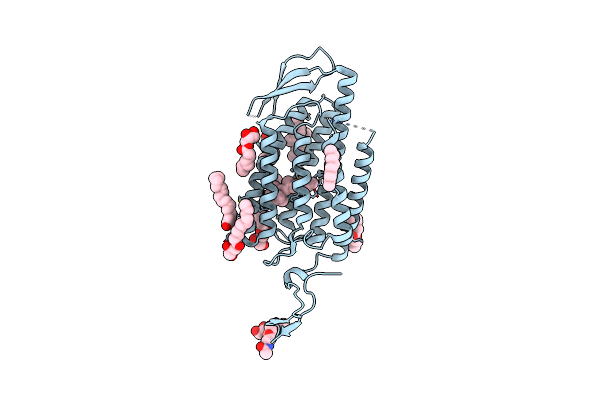
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.57 Å Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: SAL, CLR, LBN |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: Dark State Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 4 Ms Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
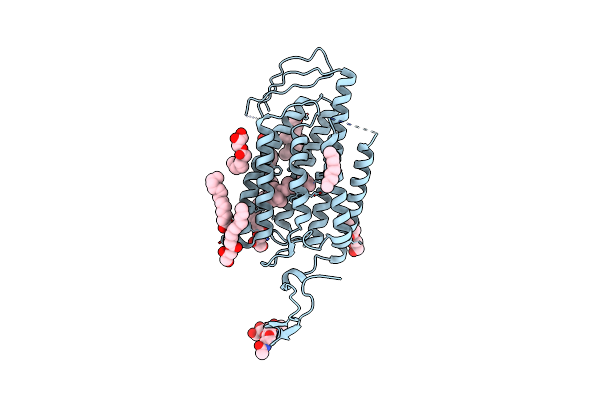
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 1 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 50 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
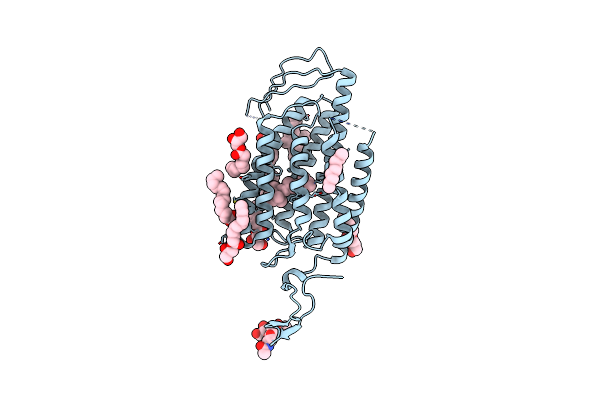
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 250 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 1 Ms Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-08-07 Classification: TRANSFERASE/IMMUNE SYSTEM |
 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3), Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-02-07 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3), Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2018-02-07 Classification: RNA BINDING PROTEIN Ligands: GTP, SO4 |

