Search Count: 48
 |
Crystal Structure Of P110Alpha-Ras Binding Domain (Rbd) In Complex With Molecular Glue D927
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: SIGNALING PROTEIN Ligands: A1ATF, MG, MRD, MPD |
 |
Crystal Structure Of P110Alpha-Ras Binding Domain (Rbd) In Complex With Molecular Glue D223
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: SIGNALING PROTEIN Ligands: A1AZD |
 |
Crystal Structure Of The Kras-P110Alpha Complex In The Presence Of Molecular Glue D223
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ONCOPROTEIN Ligands: A1AZD, GOL, MG, GNP |
 |
Crystal Structure Of Mitochondrial Citrate Synthase (Cit1) From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-04-26 Classification: TRANSFERASE |
 |
Crystal Structure Of Peroxisomal Citrate Synthase (Cit2) From Saccharomycescerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: CL, GOL, EPE, PEG |
 |
Crystal Structure Of Peroxisomal Citrate Synthase (Cit2) From Saccharomyces Cerevisiae In Complex With Oxaloacetate And Coenzyme-A
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: COA, GOL, K, CL, OAA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-04-26 Classification: TRANSFERASE/LIGASE Ligands: GOL |
 |
Crystal Structure Of F-Box Protein In The Ternary Complex With Adaptor Protein Skp1(Dl) And Its Substrate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2023-04-26 Classification: TRANSFERASE/LIGASE Ligands: EDO |
 |
Crystal Structure Of The Bacterial Oxalate Transporter Oxlt In A Ligand-Free Outward-Facing Form
Organism: Oxalobacter formigenes, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-02-15 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of The Bacterial Oxalate Transporter Oxlt In An Oxalate-Bound Occluded Form
Organism: Oxalobacter formigenes, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-02-15 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM Ligands: OXL |
 |
Crystal Structure Of Human Mmp-2 Catalytic Domain In Complex With Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-01-18 Classification: HYDROLASE Ligands: ZN, CA, 2HP, L2U |
 |
Crystal Structure Of Human Mmp-2 Catalytic Domain In Complex With Inhibitor
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-06-29 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Human Mmp-2 Catalytic Domain In Complex With Inhibitor
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-06-29 Classification: HYDROLASE Ligands: ZN, CA, B3P, CL |
 |
Organism: Drosophila melanogaster, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-01-27 Classification: CELL CYCLE Ligands: GTP, MG, SO4, GOL, GDP |
 |
Organism: Drosophila melanogaster, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.57 Å Release Date: 2021-01-27 Classification: CELL CYCLE Ligands: GTP, MG, SO4, GDP |
 |
Organism: Drosophila melanogaster, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2021-01-27 Classification: CELL CYCLE Ligands: GTP, MG, SO4, GOL, G2P |
 |
Organism: Drosophila melanogaster, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-01-27 Classification: CELL CYCLE Ligands: GTP, MG, SO4, GOL, GDP |
 |
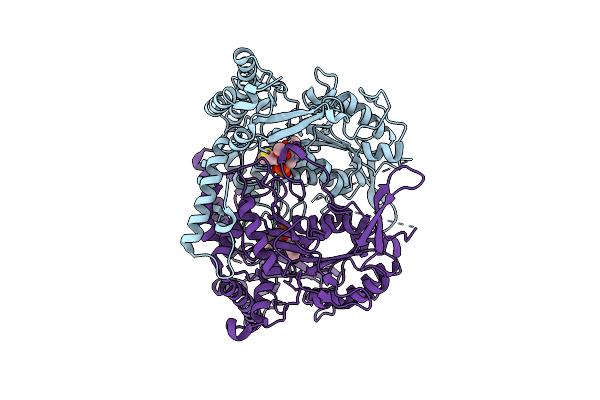
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 (Internal Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: LYASE |
 |
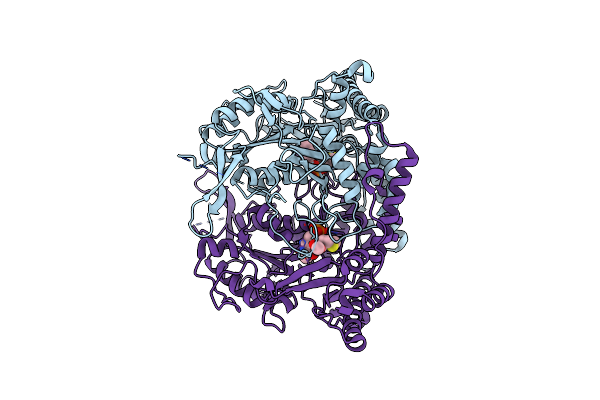
Crystal Structure Of L-Methionine Decarboxylase Q64A Mutant From Streptomyces Sp.590 In Complexed With L- Methionine Methyl Ester (Geminal Diamine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G03 |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 In Complexed With L- Methionine Methyl Ester (External Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G06 |

