Search Count: 29
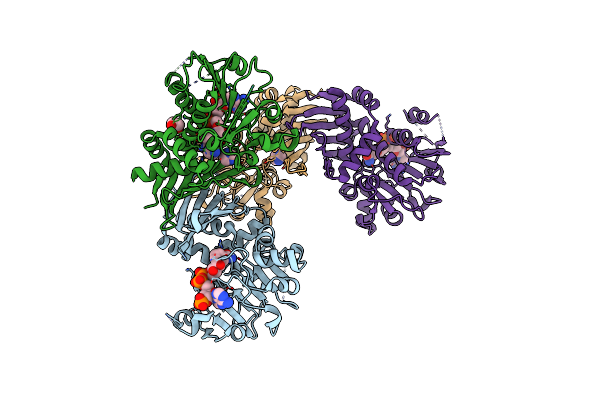 |
Organism: Pseudomonas veronii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: NAP, LYS, IMD, EDO |
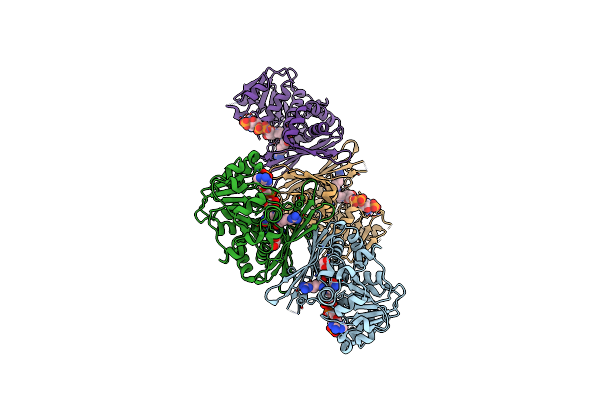 |
Organism: Pseudomonas veronii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: NDP, ARG, EDO |
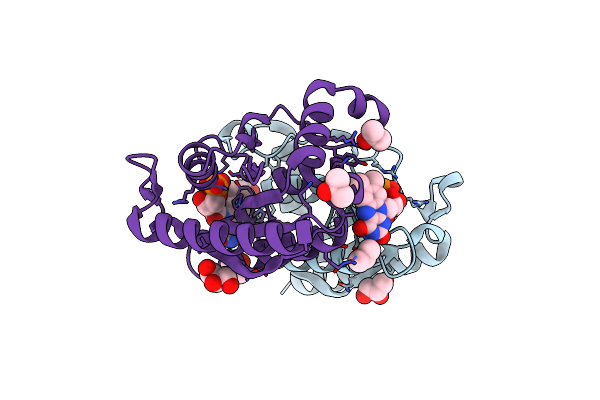 |
Crystal Structure Of Fmn-Dependent Nadph-Quinone Reductase (Azor) From Bacillus Cohnii
Organism: Bacillus cohnii
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: FMN, IPA, GOL |
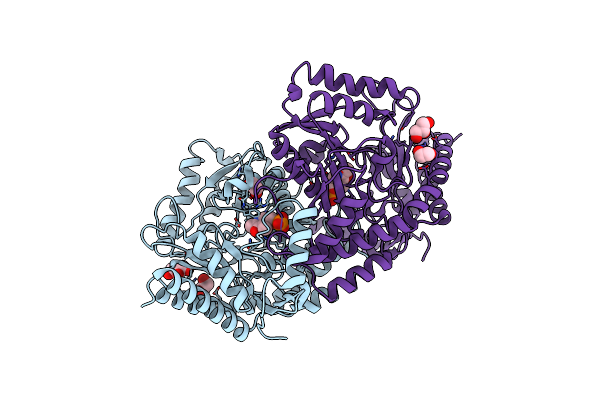 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: PLP, GOL, EDO |
 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: PLP, ORN, GOL, EDO |
 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-03-30 Classification: TRANSFERASE Ligands: PGU |
 |
Gamma-Glutamyltranspeptidase From Pseudomonas Nitroreducens Complexed With L-Don
Organism: Pseudomonas nitroreducens
Method: X-RAY DIFFRACTION Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: GOL, DON |
 |
Gamma-Glutamyltranspeptidase From Pseudomonas Nitroreducens Complexed With L-Don
Organism: Pseudomonas nitroreducens
Method: X-RAY DIFFRACTION Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: DON, GLY |
 |
Highly Active Mutant W525D Of Gamma-Glutamyltranspeptidase From Pseudomonas Nitroreducens
Organism: Pseudomonas nitroreducens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: GBL, GLY, GOL |
 |
Crystal Structure Of The Catalytic Unit Of Thermostable Gh87 Alpha-1,3-Glucanase From Streptomyces Thermodiastaticus Strain Hf3-3
Organism: Streptomyces thermodiastaticus
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2020-11-11 Classification: HYDROLASE Ligands: CA, 1PE |
 |
Catalytic Domain Of Gh87 Alpha-1,3-Glucanase From Paenibacillus Glycanilyticus Fh11
Organism: Paenibacillus glycanilyticus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, CA, GOL |
 |
Organism: Paenibacillus glycanilyticus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, CA, ACY |
 |
Catalytic Domain Of Gh87 Alpha-1,3-Glucanase D1045A In Complex With Nigerose
Organism: Paenibacillus glycanilyticus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, CA, ACY |
 |
Catalytic Domain Of Gh87 Alpha-1,3-Glucanase D1068A In Complex With Nigerose
Organism: Paenibacillus glycanilyticus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, CA, ACY |
 |
Catalytic Domain Of Gh87 Alpha-1,3-Glucanase D1069A In Complex With Nigerose
Organism: Paenibacillus glycanilyticus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, CA, ACY |
 |
Catalytic Domain Of Gh87 Alpha-1,3-Glucanase D1068A In Complex With Tetrasaccharides
Organism: Paenibacillus glycanilyticus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, CA |
 |
Catalytic Domain Of Gh87 Alpha-1,3-Glucanase D1069A In Complex With Tetrasaccharides
Organism: Paenibacillus glycanilyticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, CA |
 |
Organism: Pyrobaculum aerophilum str. im2
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2019-12-18 Classification: TRANSFERASE Ligands: ZN, FE, PO4 |
 |
Organism: Pyrobaculum aerophilum str. im2
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-12-18 Classification: TRANSFERASE Ligands: ZN, FE, ACT, UDP |
 |
Organism: Lactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-04-19 Classification: ISOMERASE Ligands: 7VO, DMS |

