Search Count: 18
 |
Crystal Structure Of Spiral2 Microtubule-Binding Domain From Physcomitrella Patens
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: PLANT PROTEIN |
 |
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-01-18 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacillus cereus (strain atcc 10987 / nrs 248)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-10-28 Classification: DNA BINDING PROTEIN |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-26 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-27 Classification: STRUCTURAL PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-27 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-08-15 Classification: REPLICATION Ligands: GDP |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-15 Classification: REPLICATION |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-08-15 Classification: REPLICATION Ligands: GDP, GSP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-08-21 Classification: STRUCTURAL PROTEIN, PROTEIN BINDING Ligands: ZN |
 |
Organism: Trachyphyllia geoffroyi
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2007-05-08 Classification: LUMINESCENT PROTEIN Ligands: NI |
 |
Organism: Trachyphyllia geoffroyi
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-05-08 Classification: LUMINESCENT PROTEIN Ligands: NI |
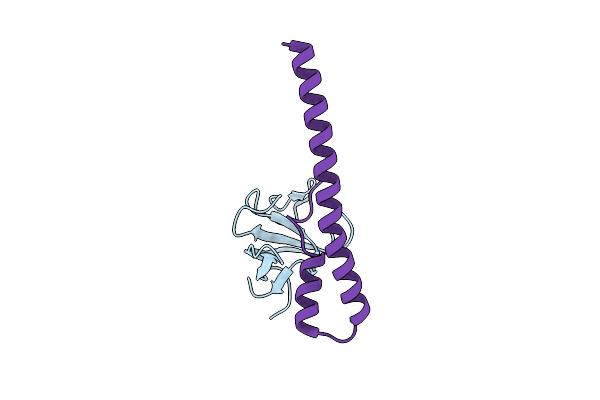 |
Crystal Structure Of The Eb1 C-Terminal Domain Complexed With The Cap-Gly Domain Of P150Glued
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-09-13 Classification: STRUCTURAL PROTEIN/PROTEIN BINDING |
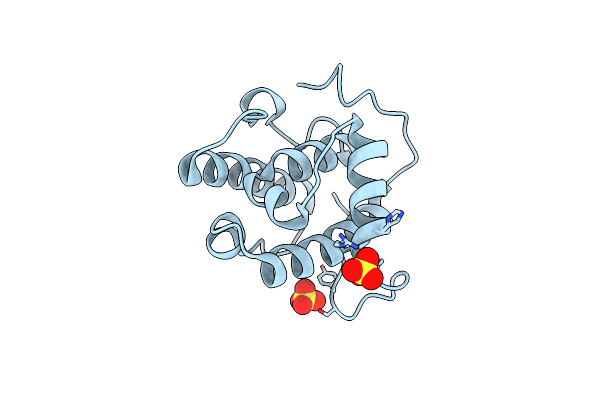 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2003-10-21 Classification: STRUCTURAL PROTEIN Ligands: SO4 |
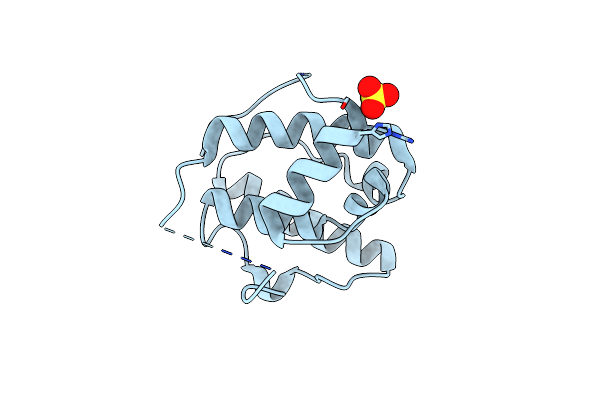 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-10-21 Classification: STRUCTURAL PROTEIN Ligands: SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2002-02-06 Classification: TRANSFERASE |
 |
Crystal Structure Analysis Of Pyrococcus Furiosus Cell Division Atpase Mind
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-04-21 Classification: cell cycle, hydrolase Ligands: MG, ADP |
 |
Crystal Structure Analysis Of Pyrococcus Furiosus Cell Division Atpase Mind
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2001-04-21 Classification: cell cycle, hydrolase Ligands: MG, ACP |

