Search Count: 192
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-277-5Cl
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7C |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-280-5I
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7A |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-276-5Br
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7B |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: PLS, HZI |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-42) (H32 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-42) (P21212 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-47) (P321 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: CA |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1Ek Complexed With F Protein Hr1 (Hr1-42) (P21 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek28 Complexed With F Protein Hr1 (Hr1-40) (H3 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MPD |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Ge Complexed With F Protein Hr1 (Hr1-42) (P21212 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: ZN, ACT |
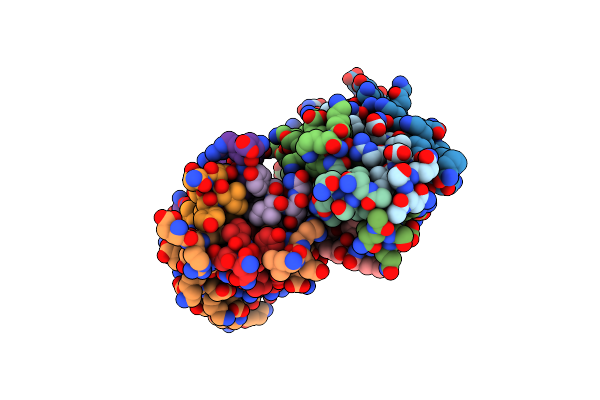 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Ge Complexed With F Protein Hr1 (Hr1-42) (P2 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
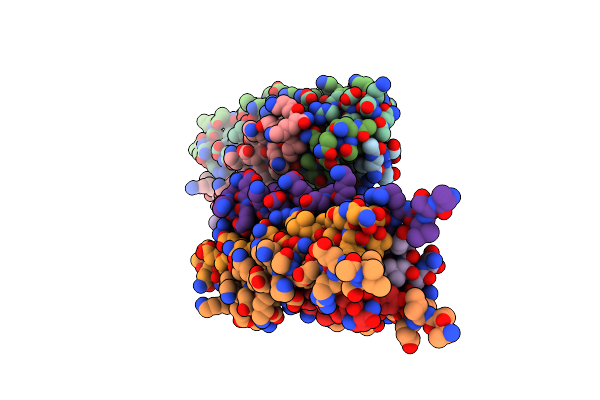 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Gt Complexed With F Protein Hr1 (Hr1-42) (P21 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PLS, A1L3N |
 |
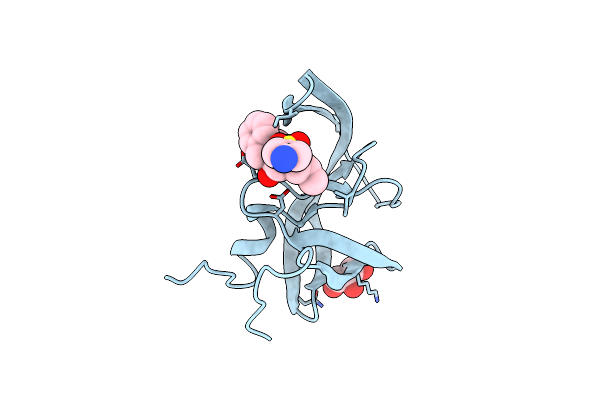
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PGE |
 |
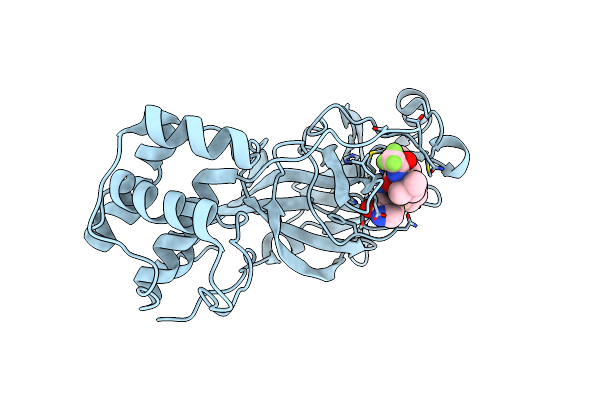
X-Ray Crystal Structure Of Multi-Drug Resistant Hiv-1 Protease (P51) In Complex With Darunavir
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN Ligands: 017, GOL |
 |
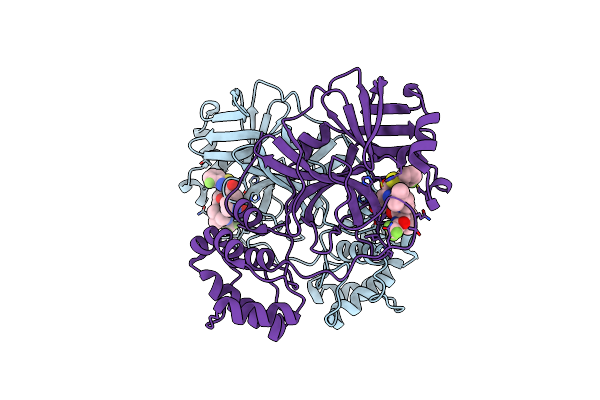
Crystal Structure Of Sars-Cov-2 Main Protease (Authentic Protein) In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: T2L |
 |
Crystal Structure Of Sars-Cov-2 Main Protease E166V Mutant In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: T2L |
 |
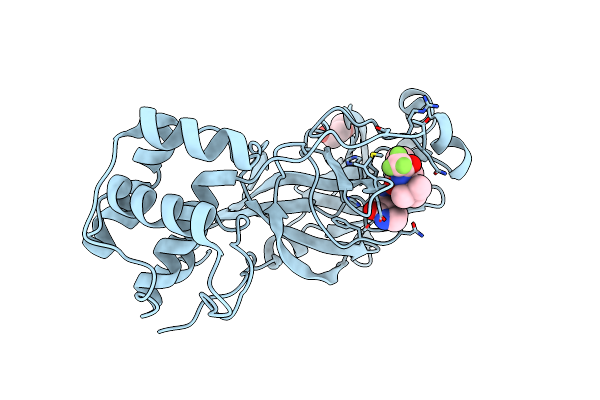
Crystal Structure Of Sars-Cov-2 Main Protease A191T Mutant In Complex With An Inhibitor 5H
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: V7G |
 |
Crystal Structure Of Sars-Cov-2 Main Protease A191T Mutant In Complex With An Inhibitor Nirmatrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI, PEG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-24 Classification: VIRAL PROTEIN |

