Search Count: 15
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-24 Classification: METAL BINDING PROTEIN Ligands: CU, GOL, ACT, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-06-24 Classification: METAL BINDING PROTEIN Ligands: CU, ZN, ACT |
 |
Crystal Structure Of Chey D13K Y106W Alone And In Complex With A Flim Peptide
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-10-05 Classification: SIGNALING PROTEIN Ligands: SO4 |
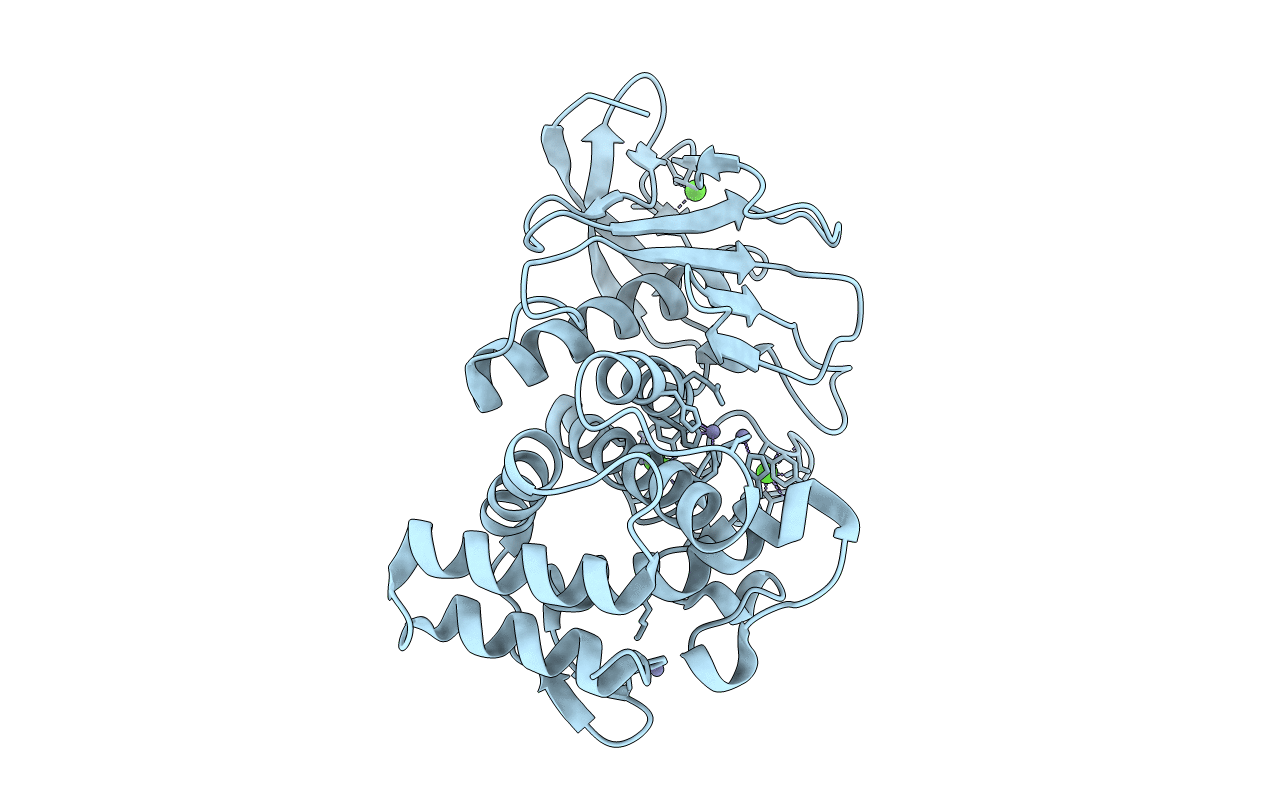 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-07-03 Classification: HYDROLASE Ligands: CA, ZN |
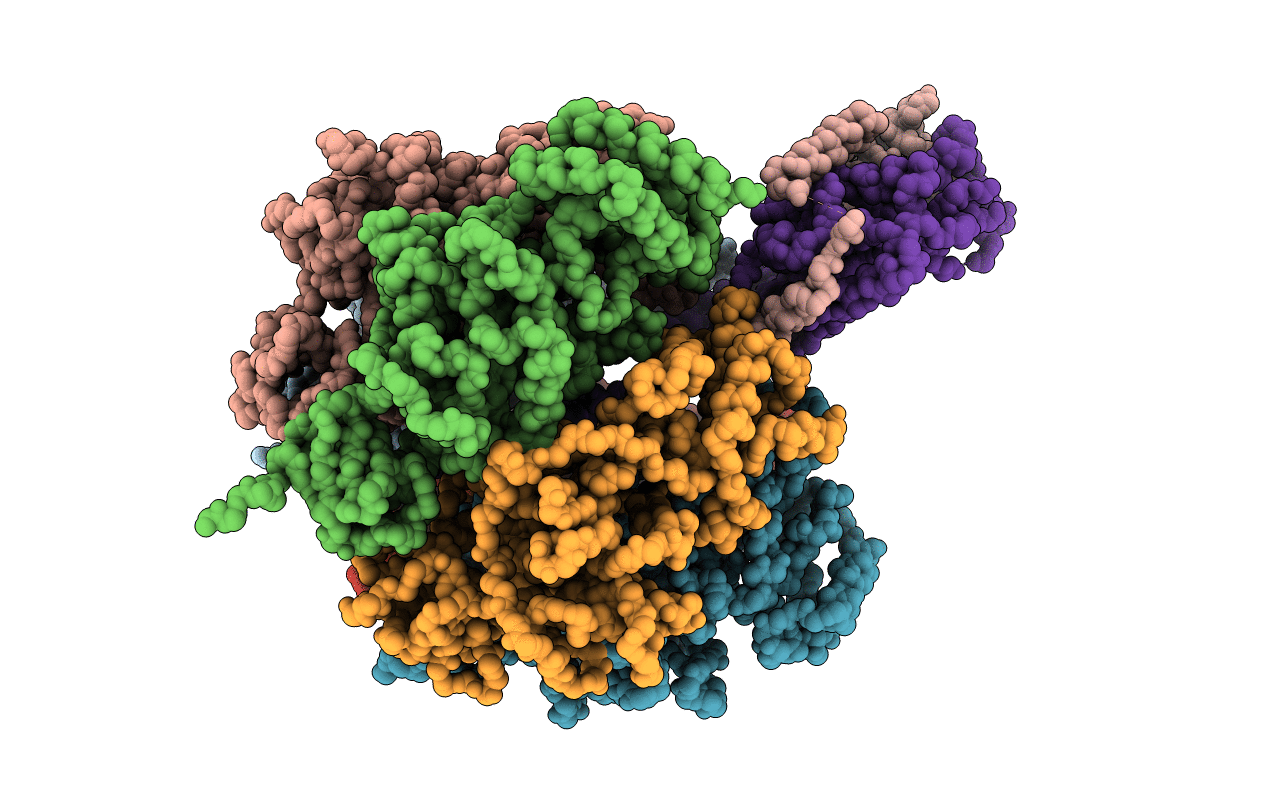 |
The Conformation Of The Epsilon And Gamma Subunits Within The E. Coli F1 Atpase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.40 Å Release Date: 2001-12-21 Classification: HYDROLASE |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 1999-12-15 Classification: HYDROLASE Ligands: CL, HED |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.40 Å Release Date: 1999-12-03 Classification: HYDROLASE |
 |
Chey-Binding (P2) Domain Of Chea In Complex With Chey From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-07-15 Classification: SIGNAL TRANSDUCTION COMPLEX |
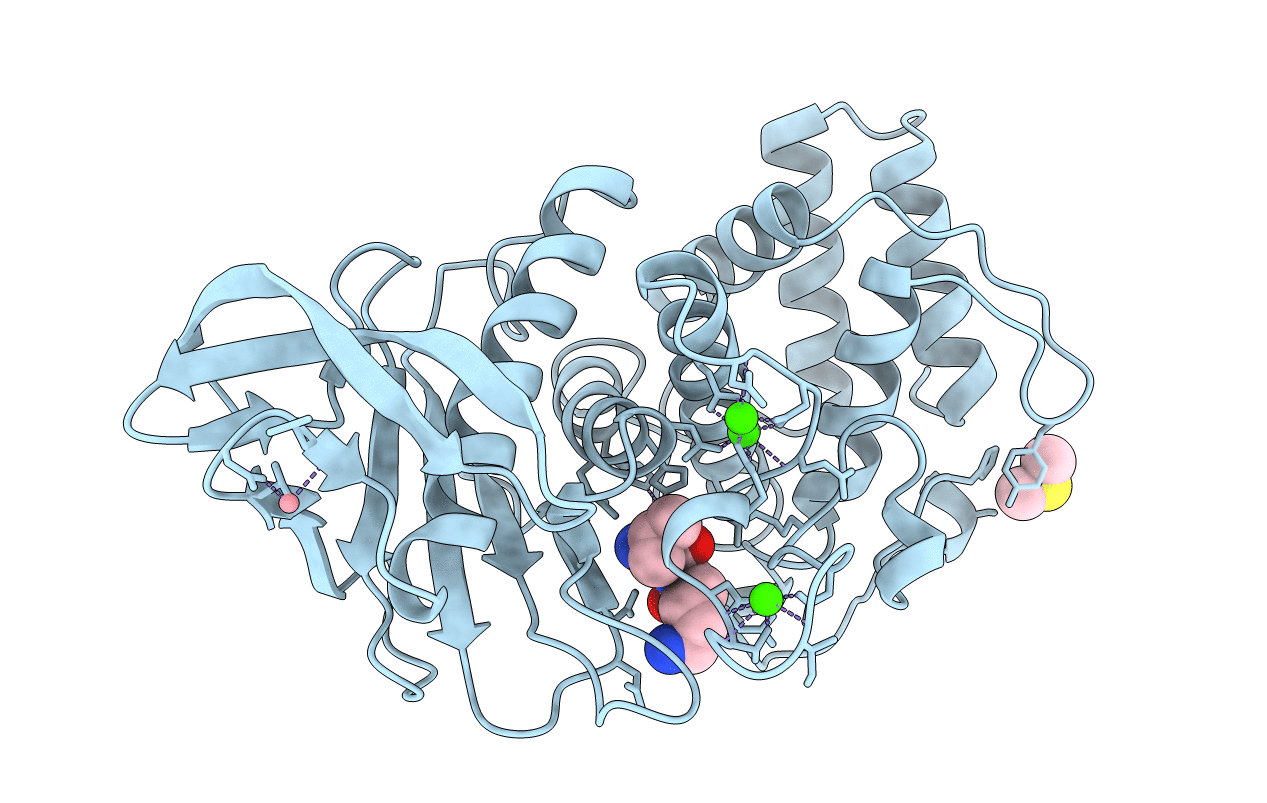 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-05-08 Classification: METALLOPROTEASE Ligands: VAL, LYS, CO, CA, DMS |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-05-08 Classification: HYDROLASE (METALLOPROTEASE) Ligands: VAL, LYS, FE, CA, DMS |
 |
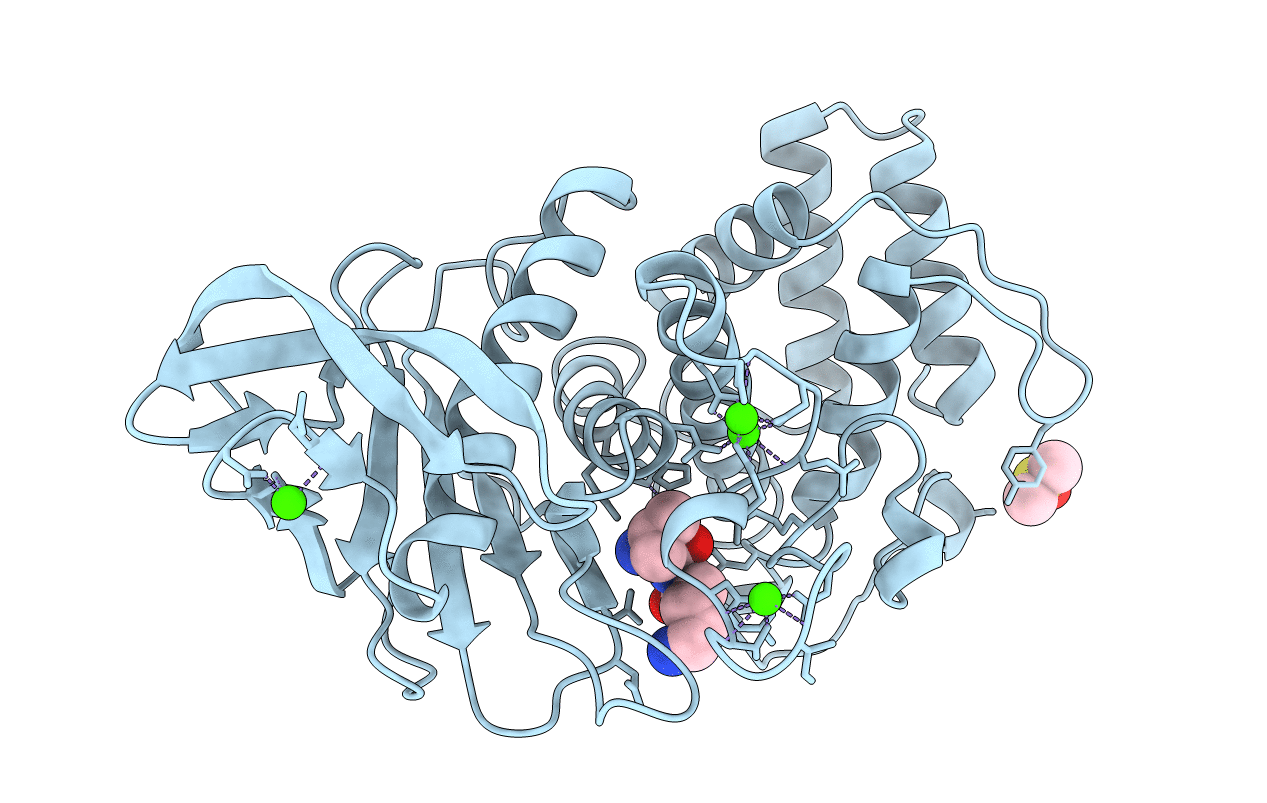
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-05-08 Classification: HYDROLASE (METALLOPROTEASE) Ligands: VAL, LYS, MN, CA, DMS |
 |
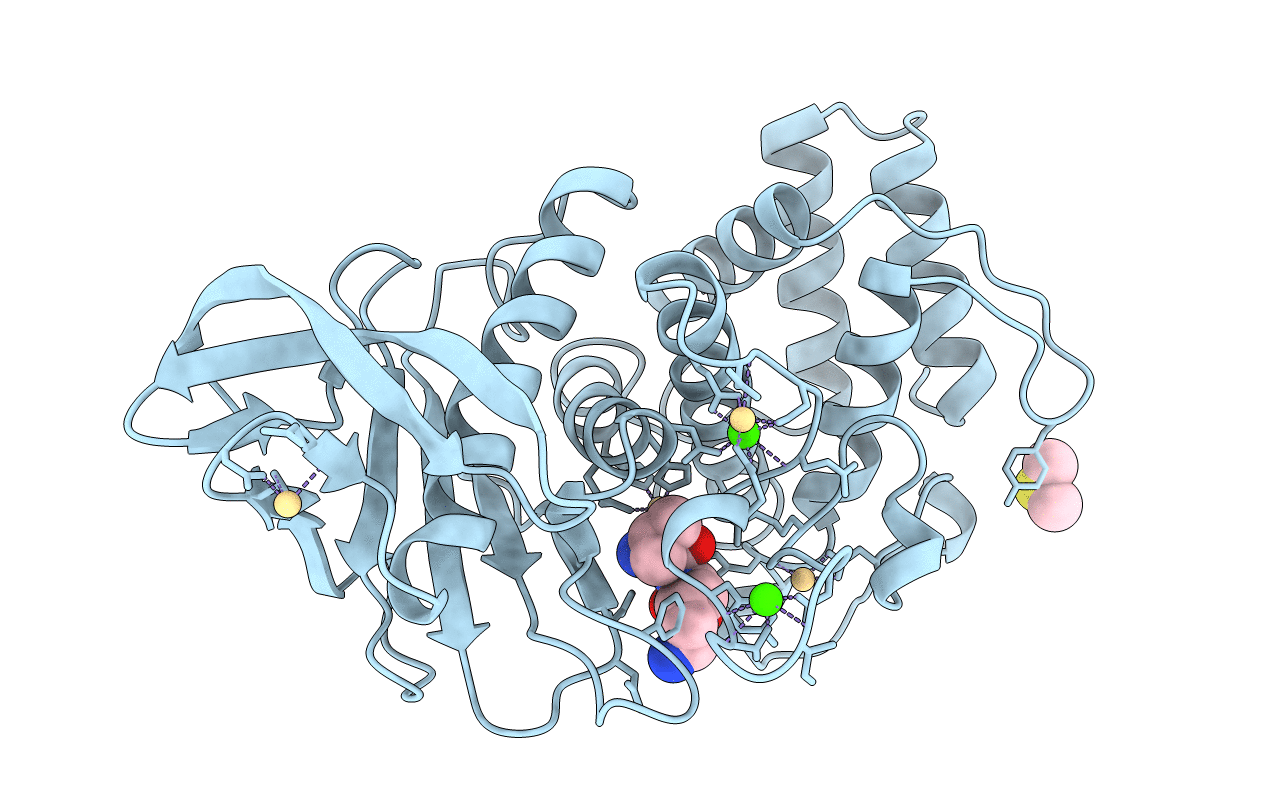
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-05-08 Classification: HYDROLASE (METALLOPROTEASE) Ligands: VAL, LYS, ZN, CA, DMS |
 |
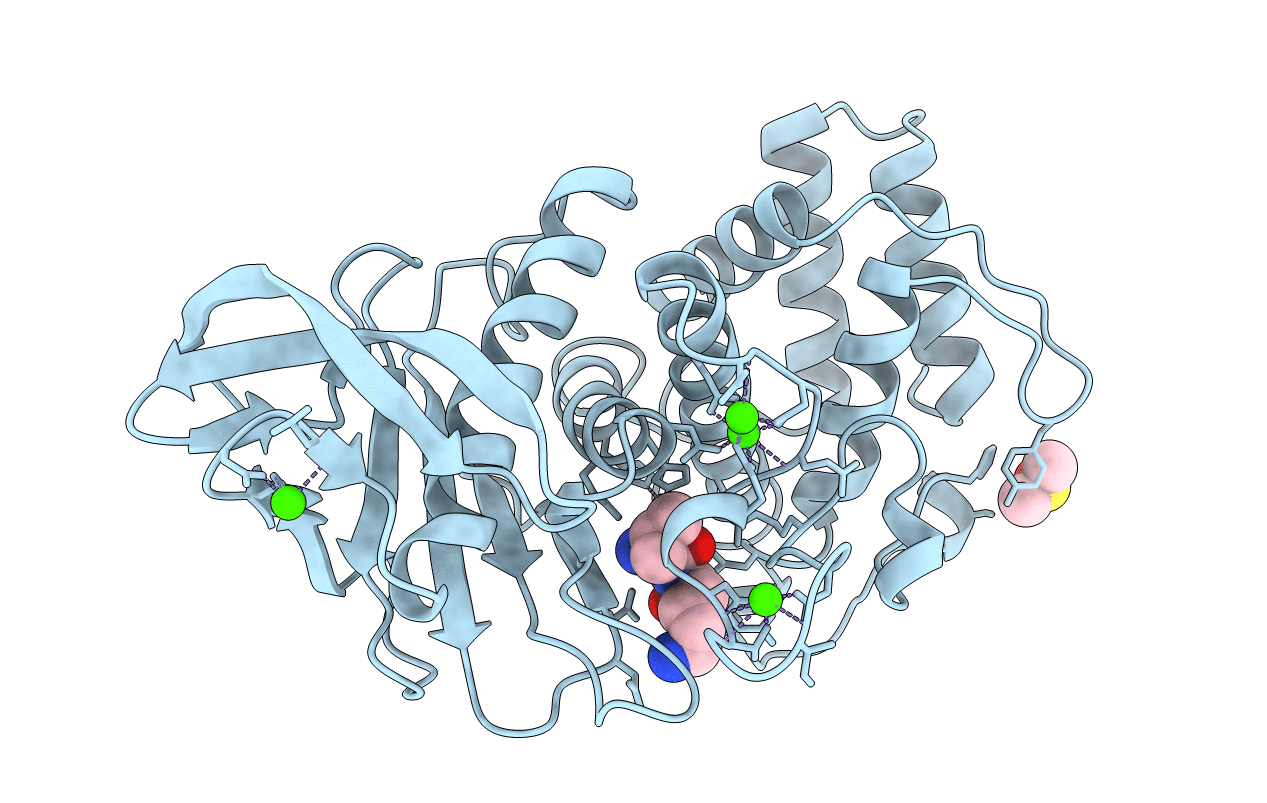
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-05-08 Classification: HYDROLASE (METALLOPROTEASE) Ligands: VAL, LYS, CD, CA, DMS |
 |
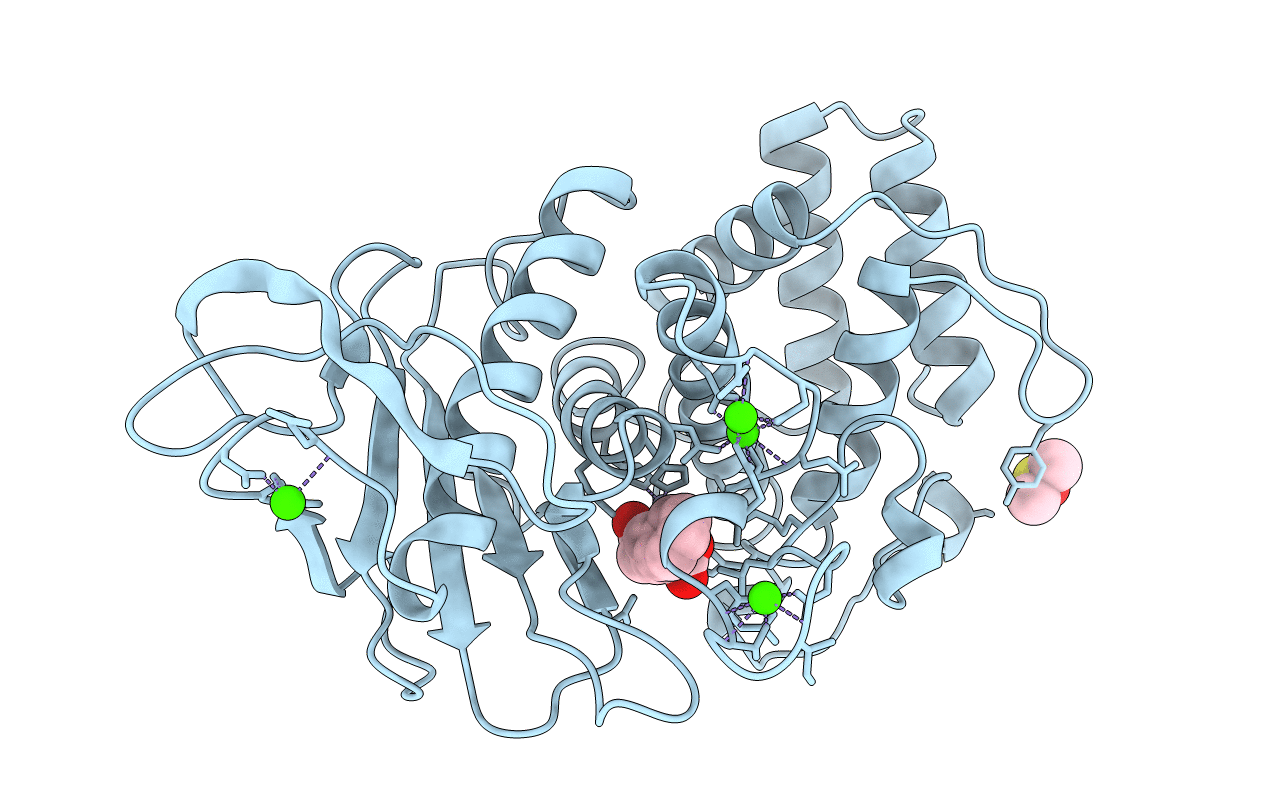
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-05-08 Classification: HYDROLASE (METALLOPROTEASE) Ligands: VAL, LYS, ZN, CA, DMS |
 |
Re-Determination And Refinement Of The Complex Of Benzylsuccinic Acid With Thermolysin And Its Relation To The Complex With Carboxypeptidase A
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-07-31 Classification: HYDROLASE(METALLOPROTEINASE) Ligands: CA, ZN, DMS, BZS |

