Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
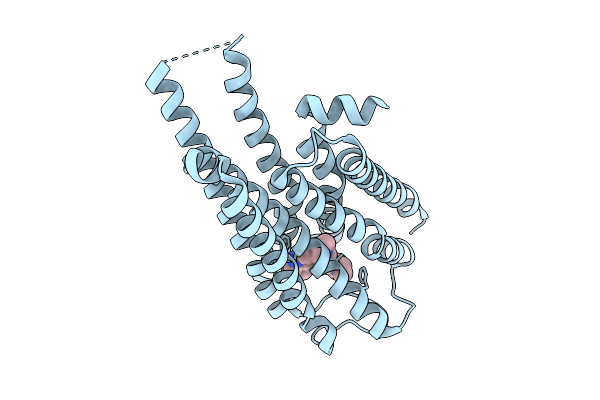 |
Local Refinement Of Drd2 Bound To Lsd In Complex With A Mini-Goa And Scfv16 Obtained By Cryo-Electron Microscopy (Cryoem)
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: MEMBRANE PROTEIN Ligands: 7LD |
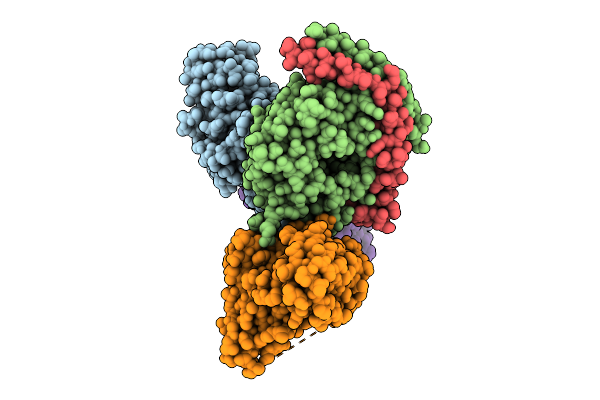 |
Global Reconstruction Of Drd2 Bound To Lsd In Complex With A Mini-Goa And Scfv16 Obtained By Cryo-Electron Microscopy (Cryoem)
Organism: Homo sapiens, Escherichia coli, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: MEMBRANE PROTEIN Ligands: 7LD |
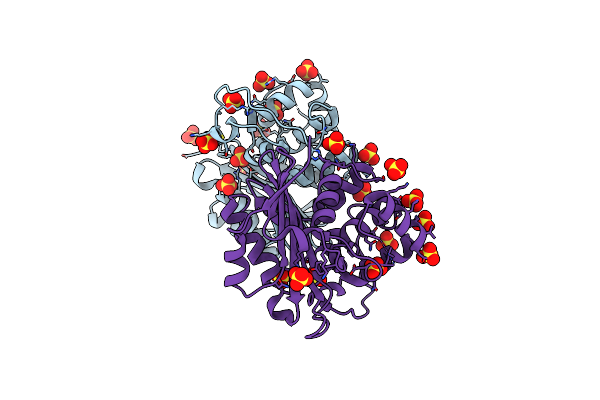 |
Crystal Structure Of The Class D Beta-Lactamase Oxa-935 From Pseudomonas Aeruginosa, Orthorhombic Crystal Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: SO4 |
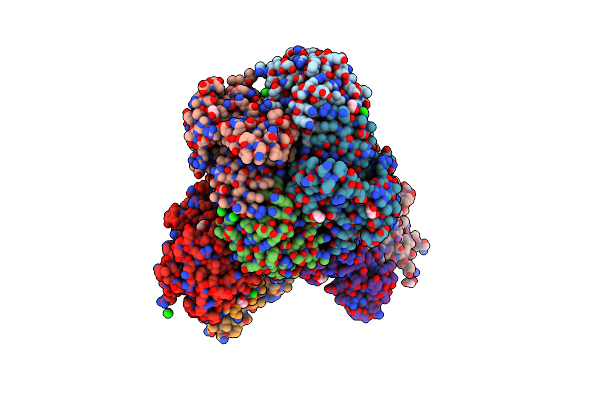 |
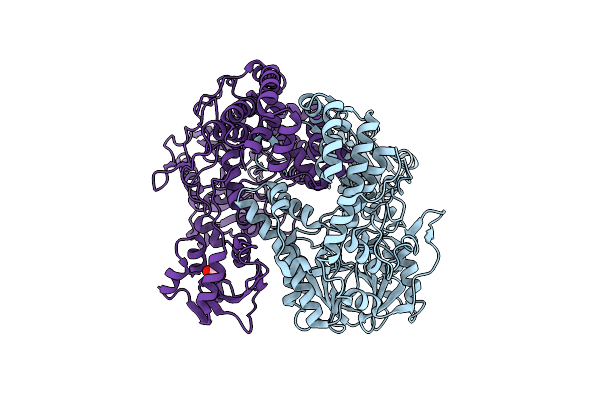
Crystal Structure Of Putataive Short-Chain Dehydrogenase/Reductase (Fabg) From Klebsiella Pneumoniae Subsp. Pneumoniae Ntuh-K2044 In Complex With Nadh
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: K, CL, NAI, GOL, EDO |
 |
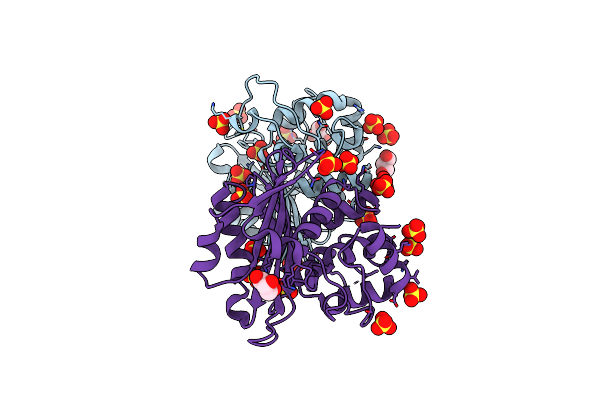
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: EDO |
 |
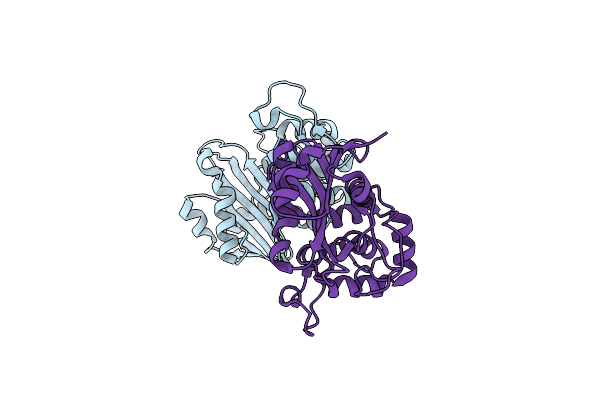
Crystal Structure Of The Oxacillin-Hydrolyzing Class D Extended-Spectrum Beta-Lactamase Oxa-14 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-12-29 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
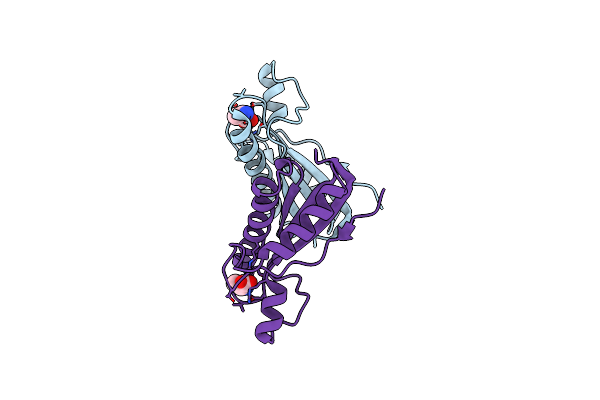
Crystal Structure Of The Class D Beta-Lactamase Oxa-935 From Pseudomonas Aeruginosa, Monoclinic Crystal Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-12-29 Classification: HYDROLASE |
 |
Crystal Structure Of The Peptidoglycan Binding Domain Of The Outer Membrane Protein (Ompa) From Klebsiella Pneumoniae With Bound D-Alanine
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-28 Classification: PEPTIDE BINDING PROTEIN Ligands: DAL, CL |
 |
The Crystal Structure Of A Functional Uncharacterized Protein Kp1_0663 From Klebsiella Pneumoniae Subsp. Pneumoniae Ntuh-K2044
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-03 Classification: UNKNOWN FUNCTION |
 |
1.52 Angstrom Resolution Crystal Structure Of Transcriptional Regulator Hdfr From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-05-06 Classification: TRANSCRIPTION Ligands: CL |
 |
2.70 Angstrom Resolution Crystal Structure Of Uracil Phosphoribosyl Transferase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-05-06 Classification: TRANSFERASE Ligands: BDF, SO4, CL |
 |
1.75 Angstrom Resolution Crystal Structure Of The Toxic C-Terminal Tip Of Cdia From Pseudomonas Aeruginosa In Complex With Immune Protein
Organism: Pseudomonas aeruginosa, Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-05-01 Classification: TOXIN |
 |
Crystal Structure Of The Sugar Binding Domain Of Laci Family Protein From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-12-26 Classification: SUGAR BINDING PROTEIN |
 |
1.55 Angstrom Resolution Crystal Structure Of 6-Phosphogluconolactonase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: B3P, CL |
 |
2.05 Angstrom Resolution Crystal Structure Of Hypothetical Protein Kp1_5497 From Klebsiella Pneumoniae.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-12-19 Classification: UNKNOWN FUNCTION Ligands: PO4, CL |
 |
1.25 Angstrom Resolution Crystal Structure Of 4-Hydroxythreonine-4-Phosphate Dehydrogenase From Klebsiella Pneumoniae.
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: NI, FMT, POP, SO4, CL |
 |
2.25 Angstrom Resolution Crystal Structure Of 6-Phospho-Alpha-Glucosidase From Klebsiella Pneumoniae In Complex With Nad And Mn2+.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-07-18 Classification: HYDROLASE Ligands: NAD, MN, SO4, CL, PEG, GOL |
 |
1.95 Angstrom Resolution Crystal Structure Of Dsba Disulfide Interchange Protein From Klebsiella Pneumoniae.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-07-11 Classification: OXIDOREDUCTASE Ligands: PGE |
 |
2.25 Angstrom Resolution Crystal Structure Of 6-Phospho-Alpha-Glucosidase From Klebsiella Pneumoniae In Complex With Nad.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-07-04 Classification: HYDROLASE Ligands: NAD, LMR, ACT, GOL, CL, BCT |
 |
1.2 Angstrom Resolution Crystal Structure Of Nucleoside Triphosphatase Nudi From Klebsiella Pneumoniae In Complex With Hepes
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2018-06-27 Classification: HYDROLASE Ligands: EPE |