Search Count: 17
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PROTEIN TRANSPORT |
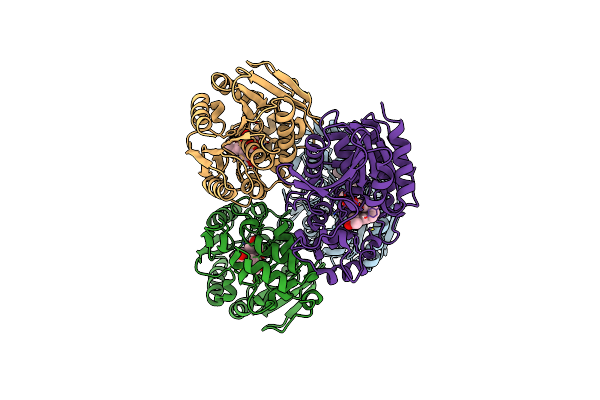 |
Zearalenone Lactonase Of Streptomyces Coelicoflavus Mutant H286Y In Complex With Hydrolyzed Zearalenone
Organism: Streptomyces coelicoflavus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2024-02-28 Classification: HYDROLASE Ligands: ZGR |
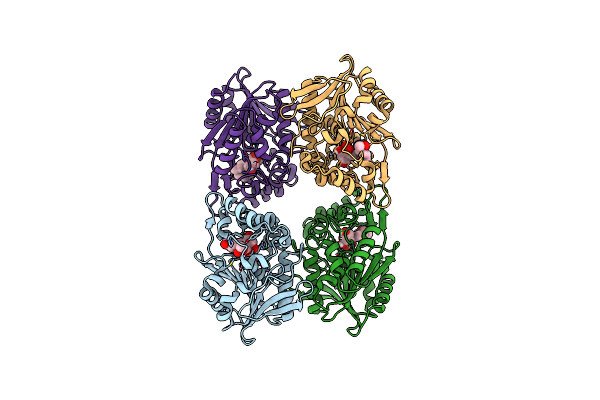 |
Zearalenone Lactonase Of Rhodococcus Erythropolis In Complex With Hydrolyzed Zearalenone
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2024-02-28 Classification: HYDROLASE Ligands: ZGR |
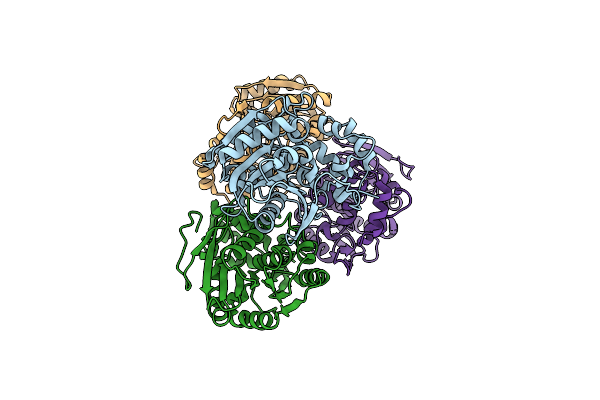 |
Zearalenone Lactonase From Streptomyces Coelicoflavus, Semet Derivative For Sad Phasing
Organism: Streptomyces coelicoflavus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-02-21 Classification: HYDROLASE |
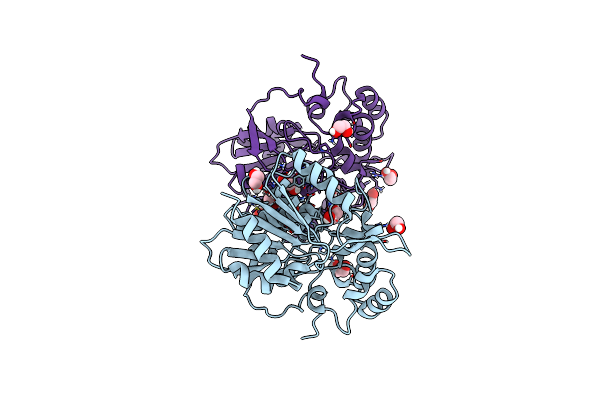 |
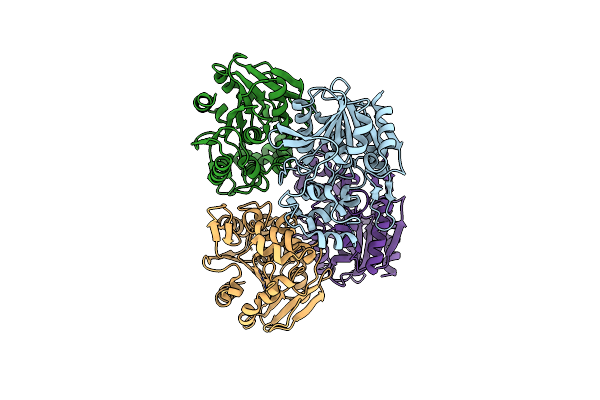
Organism: Streptomyces coelicoflavus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: EDO |
 |
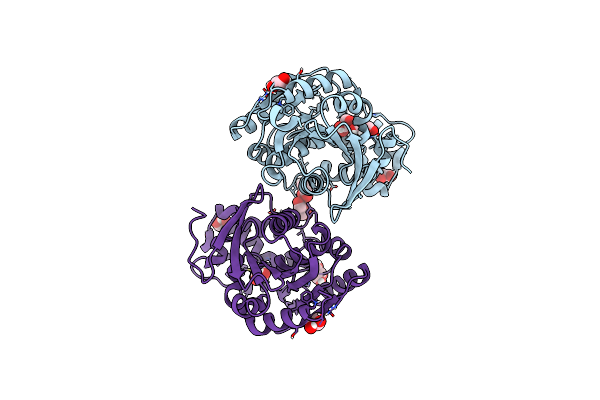
Organism: Streptomyces coelicoflavus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-02-21 Classification: HYDROLASE |
 |
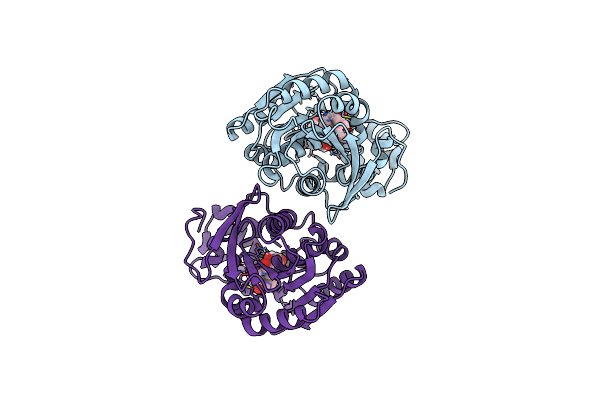
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: GOL |
 |
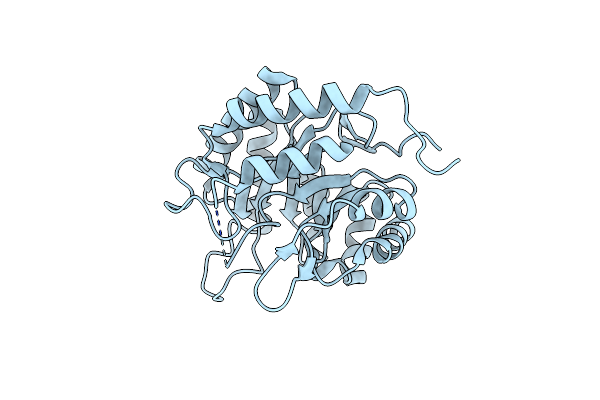
Zearalenone Lactonase From Rhodococcus Erythropolis In Complex With Zearalactamenone
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: V0F, GOL |
 |
Organism: Escherichia coli (strain b / bl21-de3)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-09-02 Classification: TRANSFERASE |
 |
Membrane Protein Megahertz Crystallography At The European Xfel, Photosystem I Xfel At 2.9 A
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-11-27 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG, CA |
 |
Membrane Protein Megahertz Crystallography At The European Xfel, Photosystem I At Synchrotron To 2.9 A
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-11-20 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG, CA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2018-10-10 Classification: HYDROLASE Ligands: CL, NA, ACT, EDO |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-10-10 Classification: ANTIBIOTIC Ligands: NXL |
 |
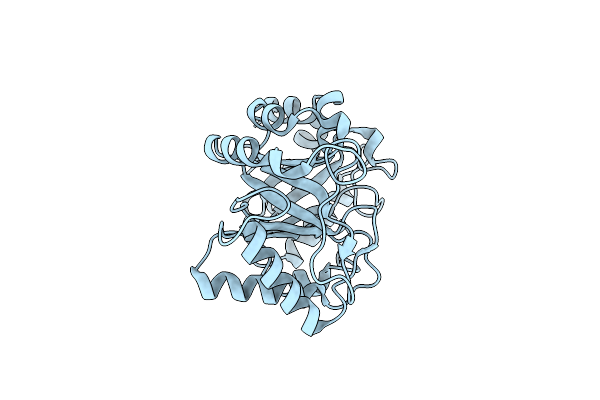
Concanavalin A Structure Determined With Data From The Euxfel, The First Mhz Free Electron Laser
Organism: Canavalia cathartica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-05 Classification: SUGAR BINDING PROTEIN Ligands: MG, CA |
 |
Concanavalin B Structure Determined With Data From The Euxfel, The First Mhz Free Electron Laser
Organism: Canavalia ensiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-05 Classification: SUGAR BINDING PROTEIN |
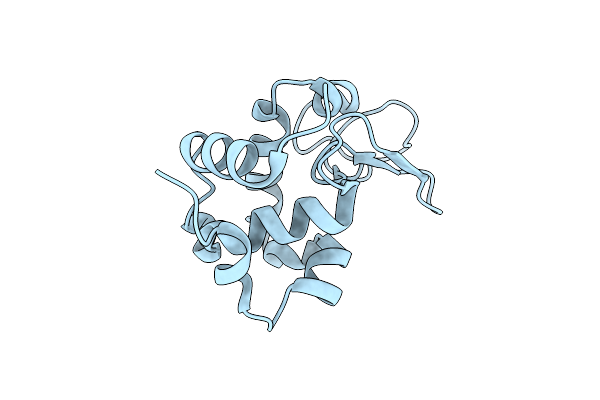 |
Hen Egg-White Lysozyme Structure Determined With Data From The Euxfel, The First Mhz Free Electron Laser, 7.47 Kev Photon Energy
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-09-05 Classification: HYDROLASE |
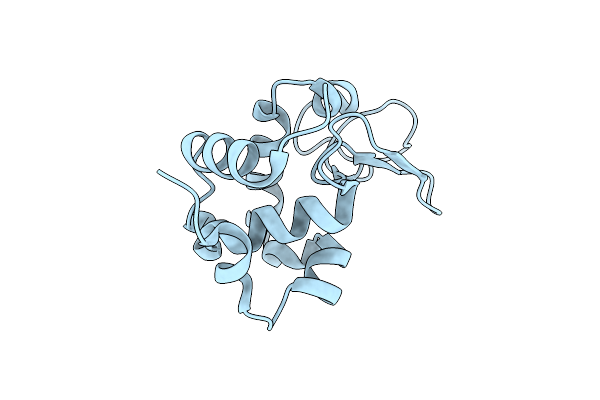 |
Hen Egg-White Lysozyme Structure Determined With Data From The Euxfel, 9.22 Kev Photon Energy
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-05 Classification: HYDROLASE |

