Search Count: 21
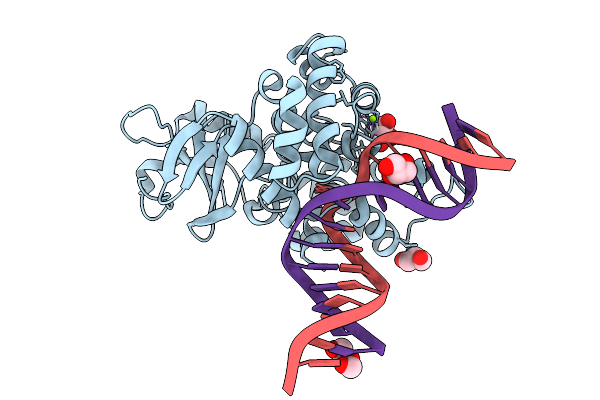 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Substrate 8-Oxoguanine Dna At Ph 8.0 Under 277 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: PEG, GOL, MG, NA |
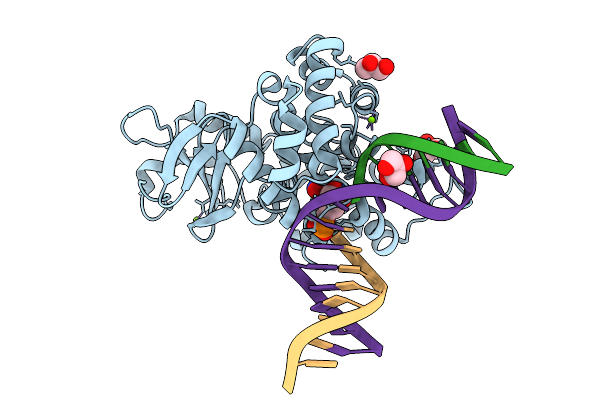 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 24 Hourss
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
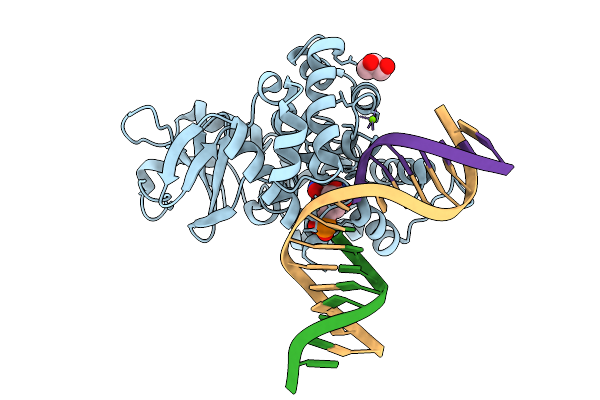 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 2.5 Hours
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
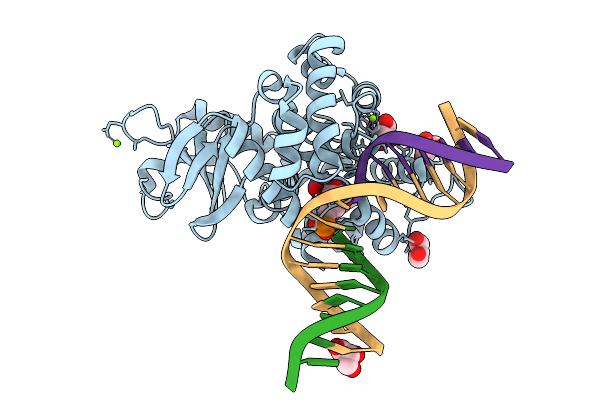 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 298 K For 3 Weeks
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, GOL, MG, NA |
 |
Crystal Structure Of Lysine Specific Demethylase 1 (Lsd1) With Tak-418 Distomer, Fad-Adduct
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-10-12 Classification: OXIDOREDUCTASE Ligands: GOL, MG, FA8, I00 |
 |
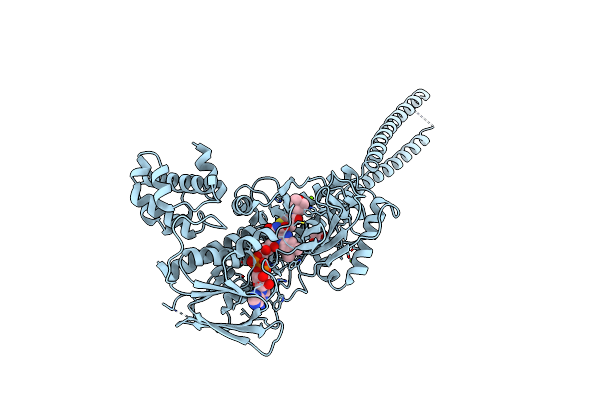
Crystal Structure Of Lysine Specific Demethylase 1 (Lsd1) With Tak-418, Fad-Adduct
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-03-24 Classification: OXIDOREDUCTASE Ligands: HUF, GOL, IMD, MG |
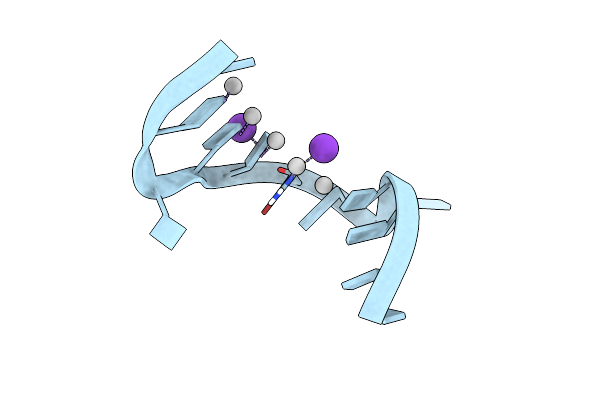 |
Crystal Structure Of Metallo-Dna Nanowire With Infinite One-Dimensional Silver Array
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-07-05 Classification: DNA Ligands: AG, K |
 |
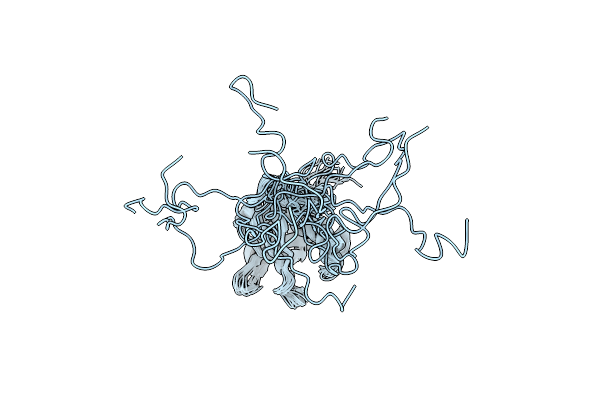 |
Solution Structure Of Mtsl Spin-Labeled Schizosaccharomyces Pombe Sin1 Crim Domain
Organism: Schizosaccharomyces pombe 972h-
Method: SOLUTION NMR Release Date: 2015-07-29 Classification: PROTEIN BINDING |
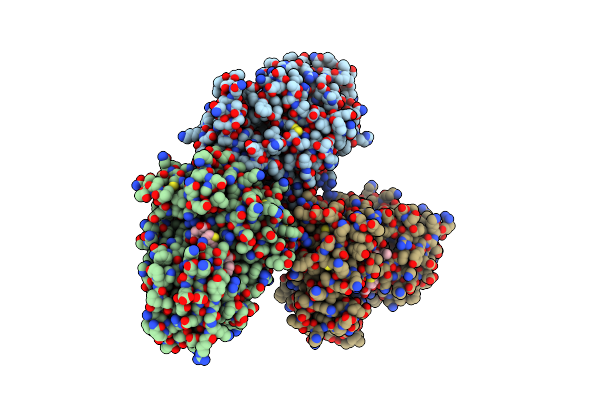 |
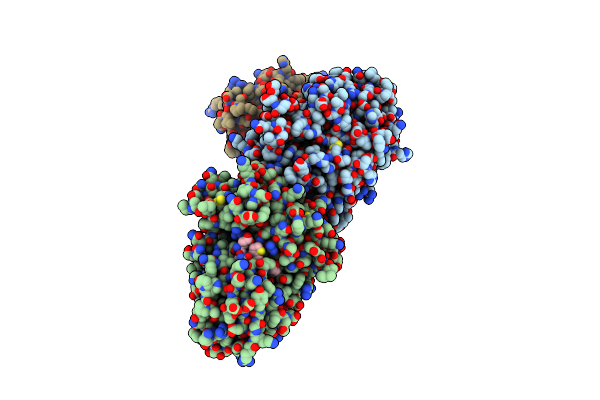
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-07-01 Classification: Hydrolase/Hydrolase inhibitor |
 |
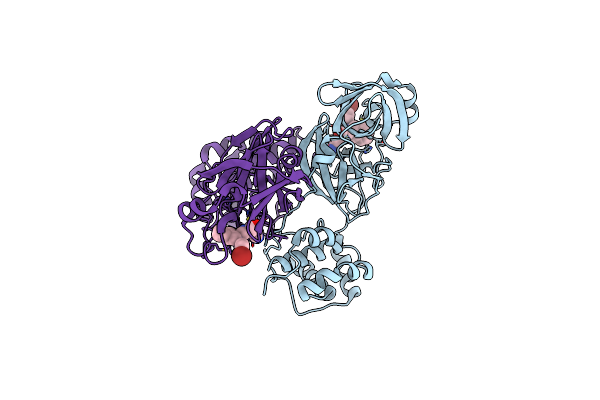
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2015-07-01 Classification: Hydrolase/Hydrolase inhibitor |
 |
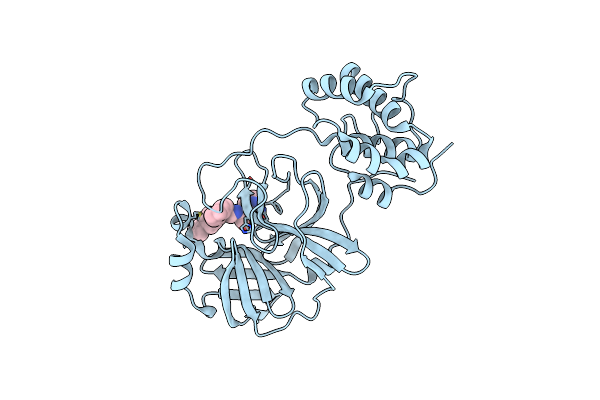
Structure Of Sars-3Cl Protease Complex With A Bromobenzoyl (S,R)-N-Decalin Type Inhibitor
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2015-02-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3A7 |
 |
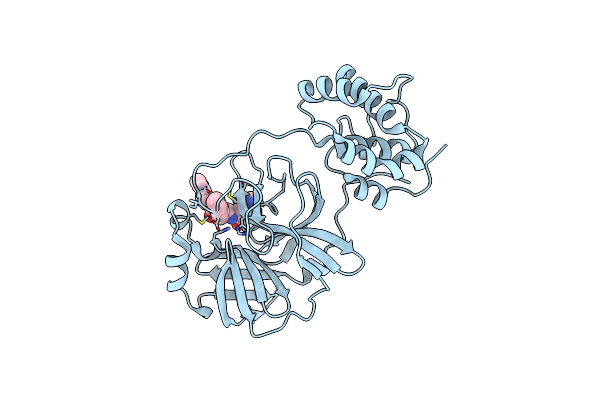
Structure Of Sars-3Cl Protease Complex With A Phenylbenzoyl (S,R)-N-Decalin Type Inhibitor
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-02-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3BL |
 |
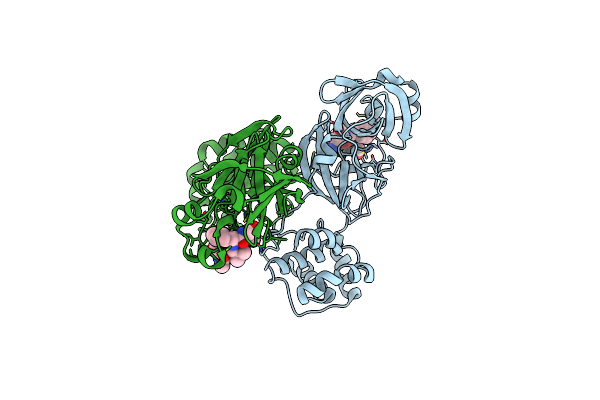
Structure Of Sars-3Cl Protease Complex With A Phenylbenzoyl (R,S)-N-Decalin Type Inhibitor
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2015-02-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3X5 |
 |
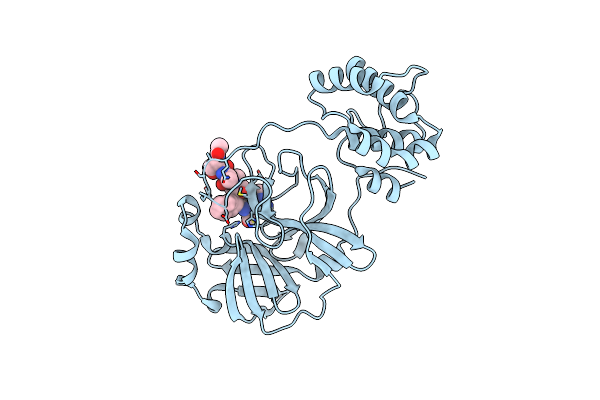
Structure-Based Design, Synthesis, Evaluation Of Peptide-Mimetic Sars 3Cl Protease Inhibitors
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2011-12-14 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
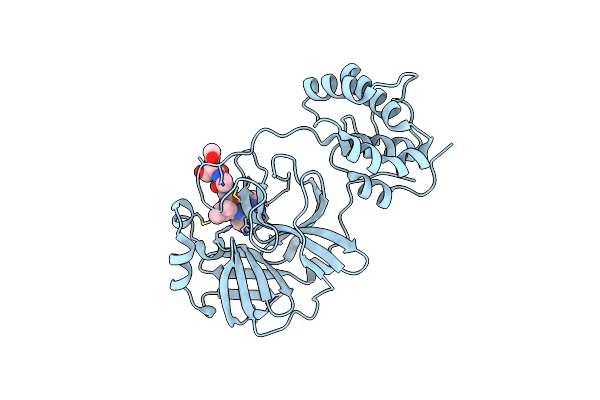
Structure Of Sars 3Cl Protease With Peptidic Aldehyde Inhibitor Containing Cyclohexyl Side Chain
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2011-12-14 Classification: HYDROLASE/HYDROLASE inhibitor |
 |
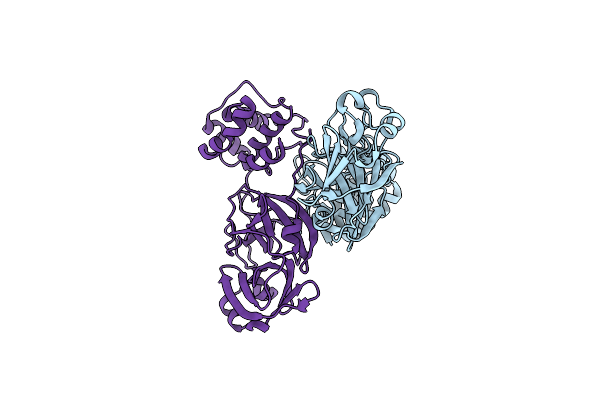
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-12-14 Classification: HYDROLASE/HYDROLASE inhibitor |
 |
Structure Of Sars 3Cl Protease Auto-Proteolysis Resistant Mutant In The Absent Of Inhibitor
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-12-14 Classification: HYDROLASE |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-07-14 Classification: ELECTRON TRANSPORT Ligands: HEC, PG4, PEG, PO4 |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-07-14 Classification: ELECTRON TRANSPORT Ligands: HEC, PG4, PEG, PGE |

