Search Count: 13
 |
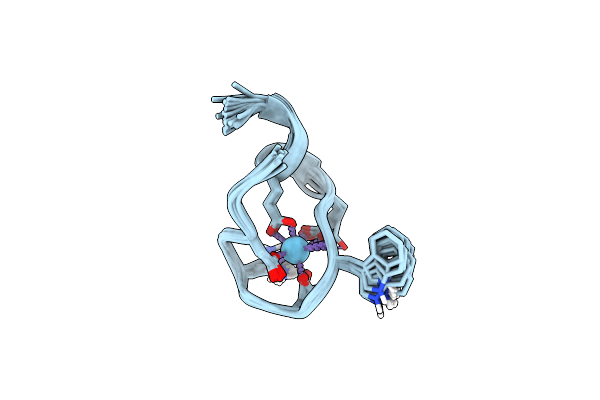
The Binding Structure Of A Lanthanide Binding Tag (Lbt3) With Lutetium Ion (Lu3+)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2021-04-28 Classification: DE NOVO PROTEIN Ligands: LU |
 |
The Binding Structure Of A Lanthanide Binding Tag (Lbt3) With Lanthanum Ion (La3+)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2021-04-28 Classification: DE NOVO PROTEIN Ligands: LA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-02-13 Classification: IMMUNE SYSTEM Ligands: CL |
 |
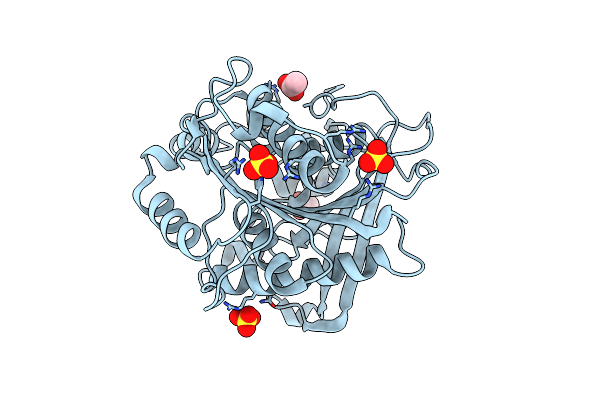
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-02-13 Classification: IMMUNE SYSTEM |
 |
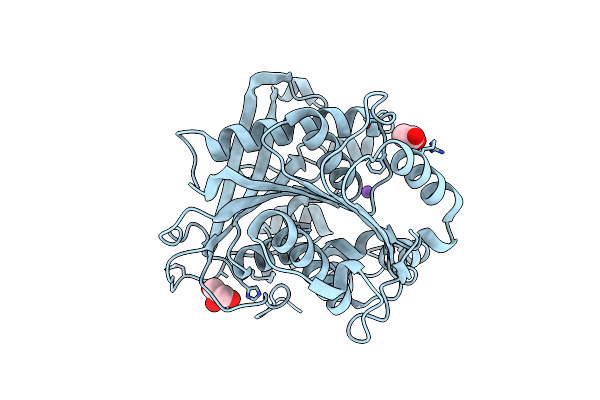
Organism: Streptomyces cinnamoneus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-12-06 Classification: HYDROLASE Ligands: SO4, ACT |
 |
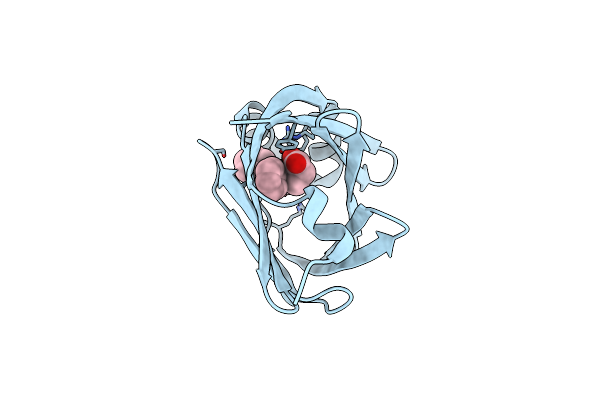
Organism: Streptomyces cinnamoneus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
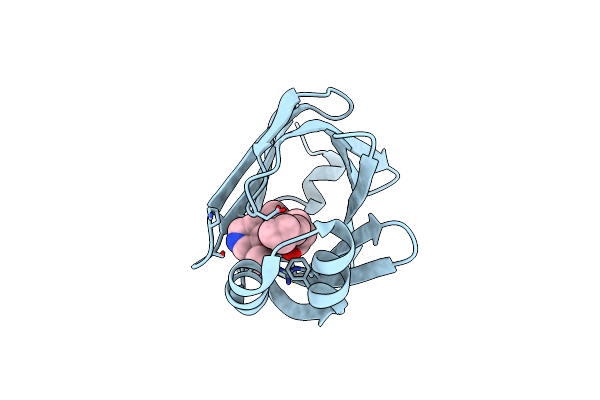
Crystal Structure Of Fabp4 In Complex With 3-(5-Cyclopropyl-2,3-Diphenyl-1H-Indol-1-Yl)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: 57P |
 |
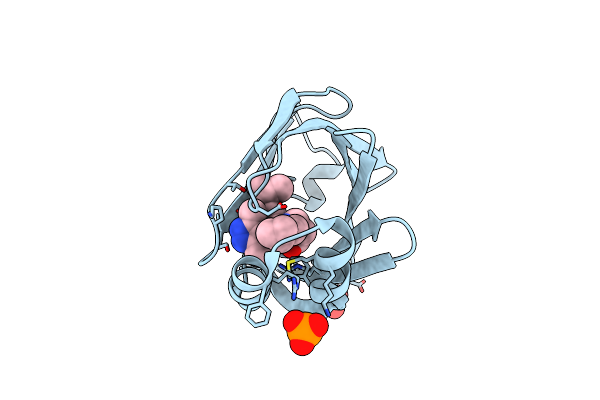
Crystal Structure Of Fabp4 In Complex With 3-[5-Cyclopropyl-3-(3-Methoxypyridin-4-Yl)-2-Phenyl-1H-Indol-1-Yl] Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: L19 |
 |
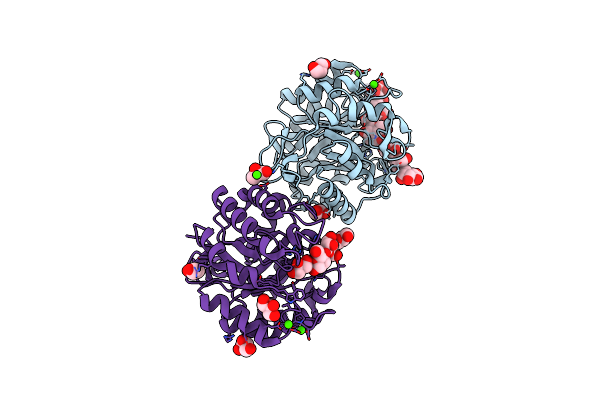
Crystal Structure Of Fabp4 In Complex With 3-{5-Cyclopropyl-3-(3,5-Dimethyl-1H-Pyrazol-4-Yl)-2-[3-(Propan-2-Yloxy) Phenyl]-1H-Indol-1-Yl}Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: L96, PO4 |
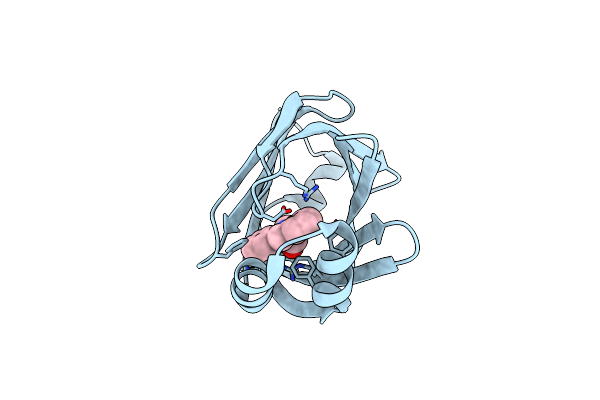 |
Crystal Structure Of Fabp4 In Complex With 3-(2-Phenyl-1H-Indol-1-Yl)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: 57Q |
 |
Crystal Structure Of Beta-Mannanase From Streptomyces Thermolilacinus With Mannohexaose
Organism: Streptomyces thermolilacinus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-09-23 Classification: HYDROLASE Ligands: GOL, BMA, CA |
 |
Organism: Streptomyces thermolilacinus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A New Alkaline Serine Protease (M-Protease) From Bacillus Sp. Ksm-K16
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-06-22 Classification: SERINE PROTEINASE Ligands: CA |

