Search Count: 14
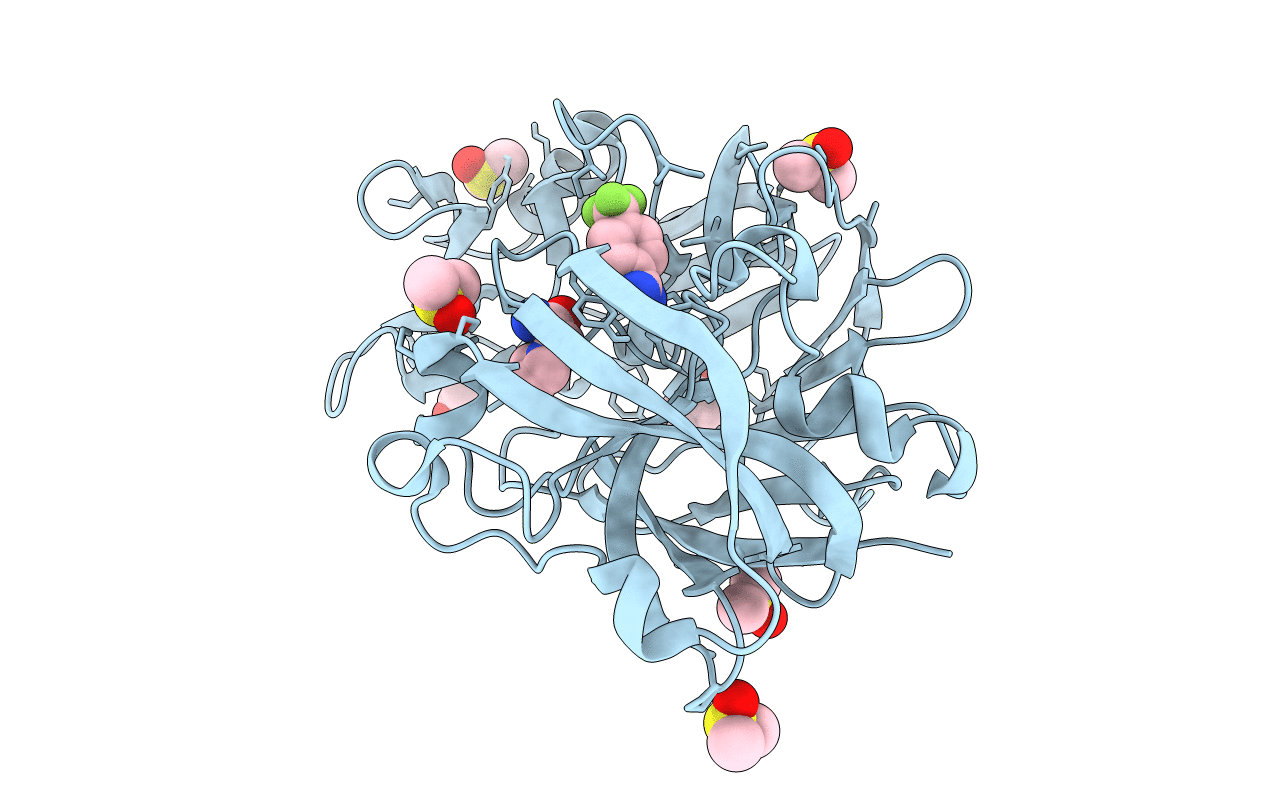 |
Cocktail Experiment G: Fragments 216 And 338 At 90 Mm Concentration In Complex With Endothiapepsin
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 483, 8NE, GOL, DMS, ACT |
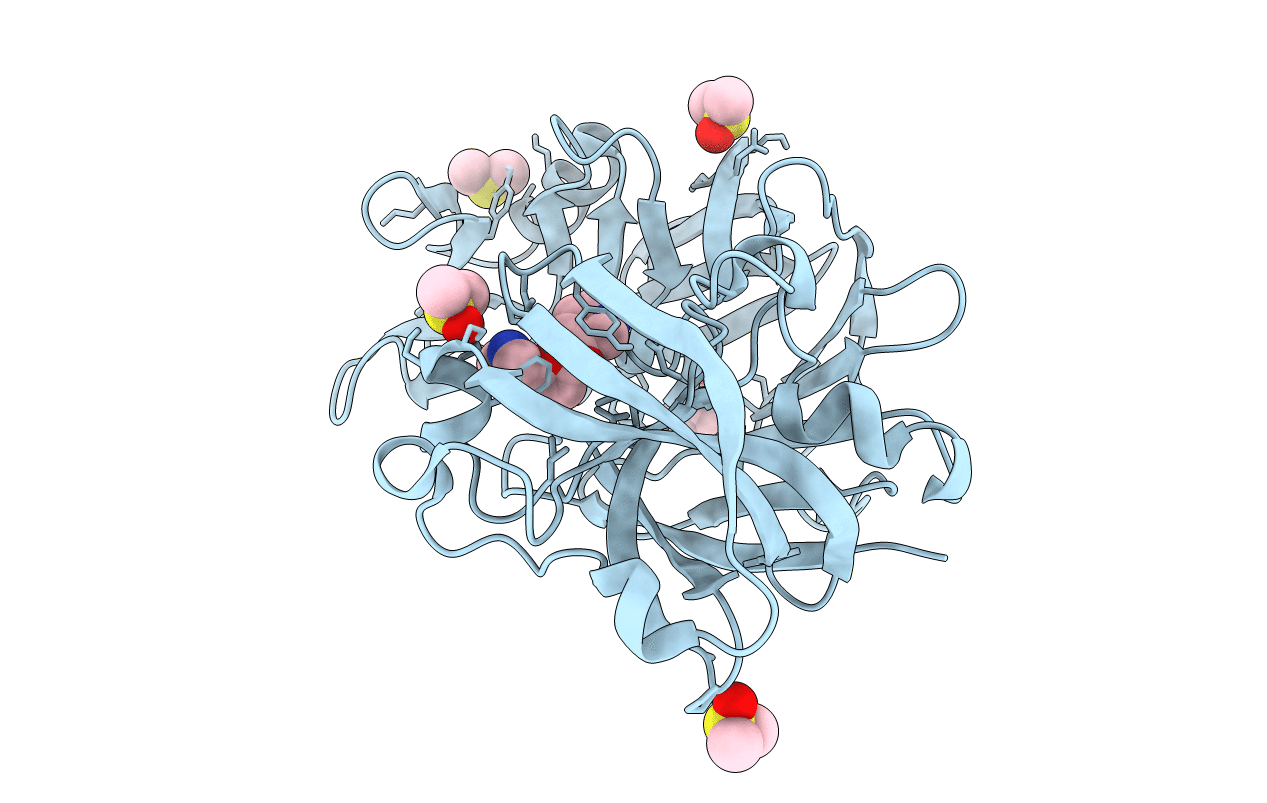 |
Cocktail Experiment E: Fragments 52, 58, And 63 At 90 Mm Concentration In Complex With Endothiapepsin
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: F63, DMS, GOL |
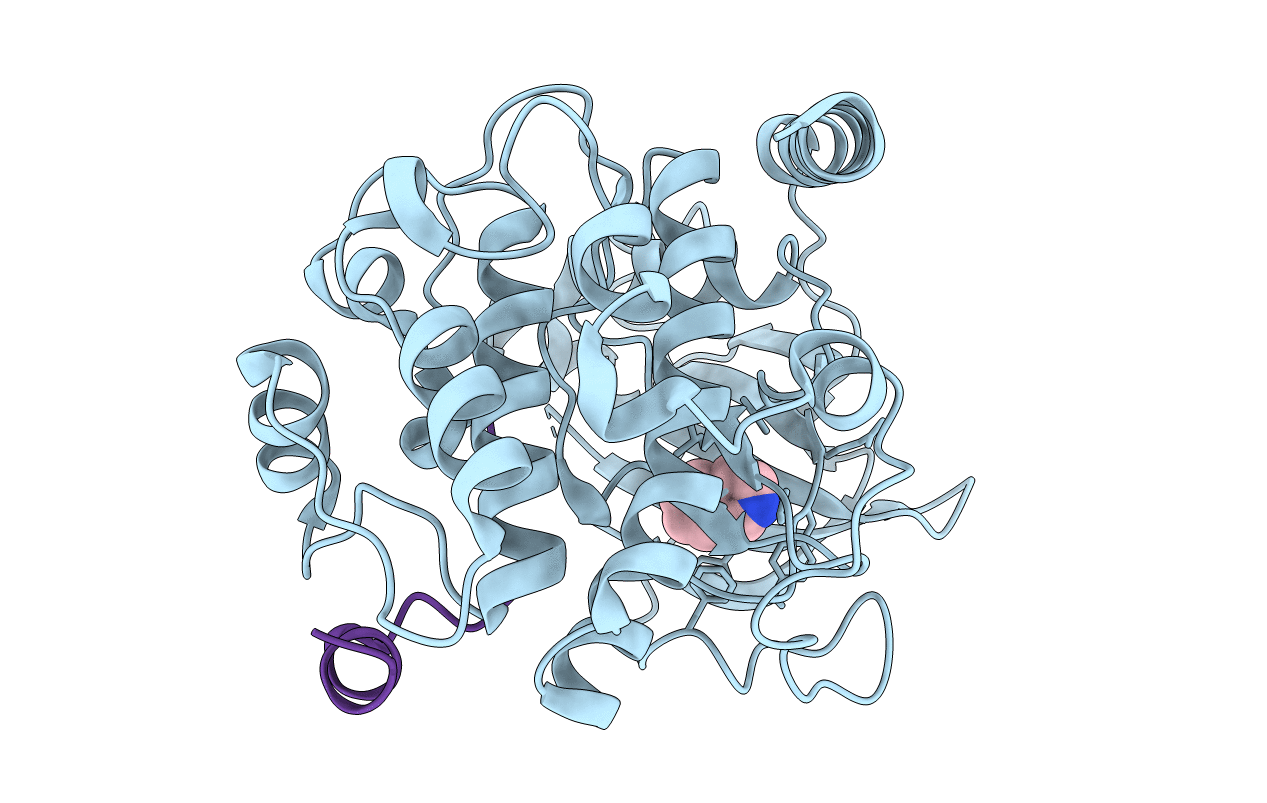 |
Crystal Structure Of The Camp-Dependent Protein Kinase A Cocrystallized With Pki (5-24). Soaking Of Aminofasudil And Displacing It With The Fragment Isoquinoline.
Organism: Cricetulus griseus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-04-28 Classification: TRANSFERASE Ligands: ISQ |
 |
Crystal Structure Of The N-Terminal Domain Of The Wild Type Parasitic Pex14
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2020-08-26 Classification: SIGNALING PROTEIN Ligands: PEG |
 |
First Crystal Structure Of Parasitic Pex14 In Complex With A Fragment Molecule 1H-Indole-7-Carboxylic Acid
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-07-15 Classification: SIGNALING PROTEIN Ligands: SO4, KXQ, GOL, DMS |
 |
Structure Of Parasitic Pex14 In Complex With A Benzo[B]Thiophene-7-Carboxylic Acid.
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-07-15 Classification: SIGNALING PROTEIN Ligands: KZ2, PEG |
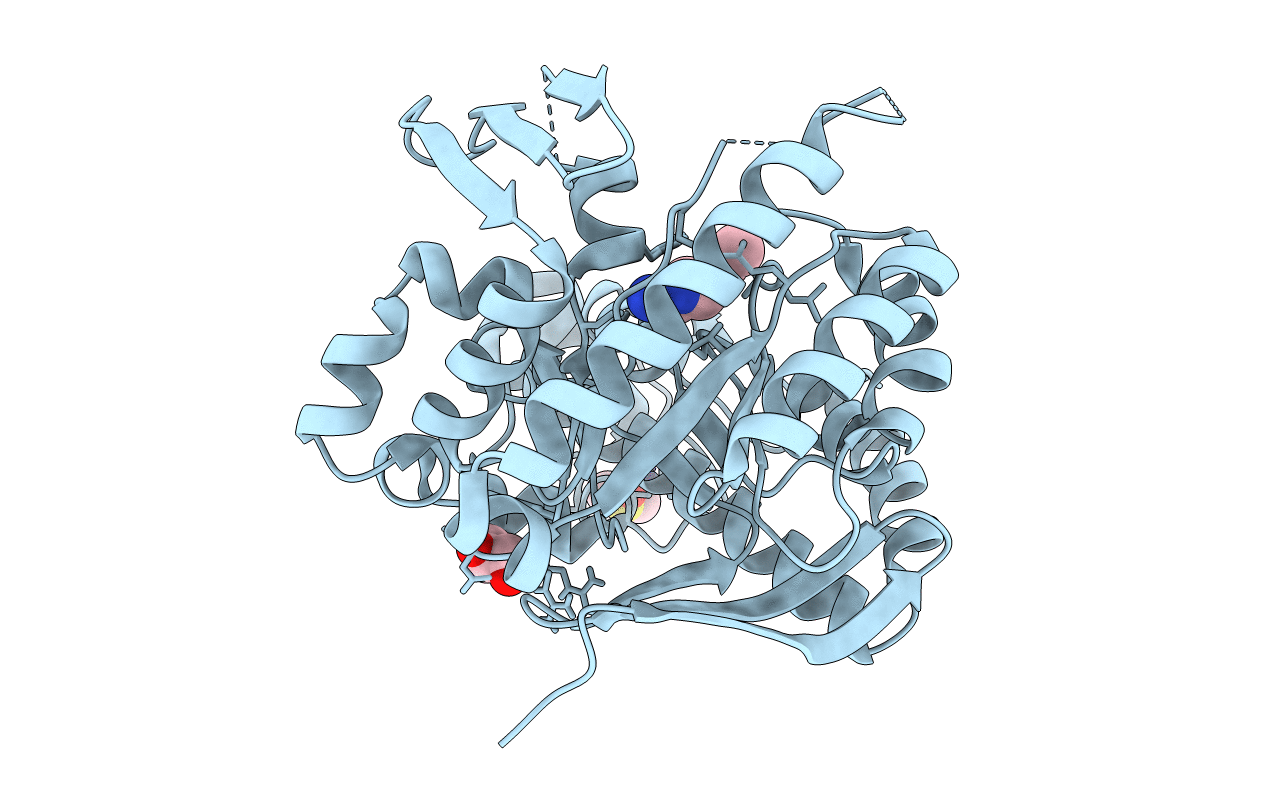 |
Crystal Structure Of Tgt In Complex With N2-Methyl-8-(Prop-1-Yn-1-Yl)-3H,7H,8H-Imidazo[4,5-G]Quinazoline-2,6-Diamine
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2020-06-03 Classification: TRANSFERASE Ligands: ZN, K75, GOL, DMS |
 |
Crystal Structure Of Tgt In Complex With N2-Methyl-1H,7H,8H-Imidazo[4,5-G]Quinazoline-2,6-Diamine
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-06-03 Classification: TRANSFERASE Ligands: GOL, DMS, ZN, K7H |
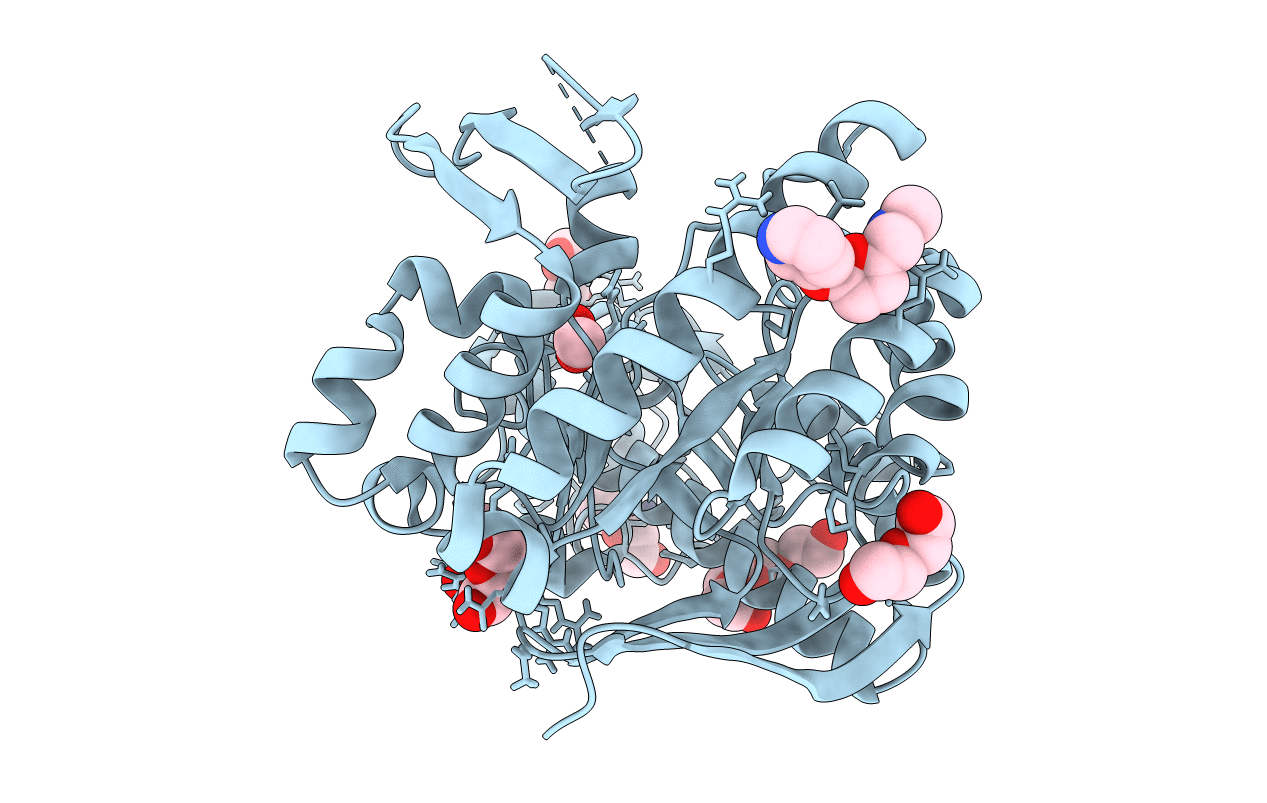 |
Crystal Structure Of Tgt In Complex With Methyl({[5-(Pyridin-3-Yloxy)Furan-2-Yl]Methyl})Amine
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-03-20 Classification: TRANSFERASE Ligands: ZN, PEG, DMS, F63, PG4, ACT, PGE |
 |
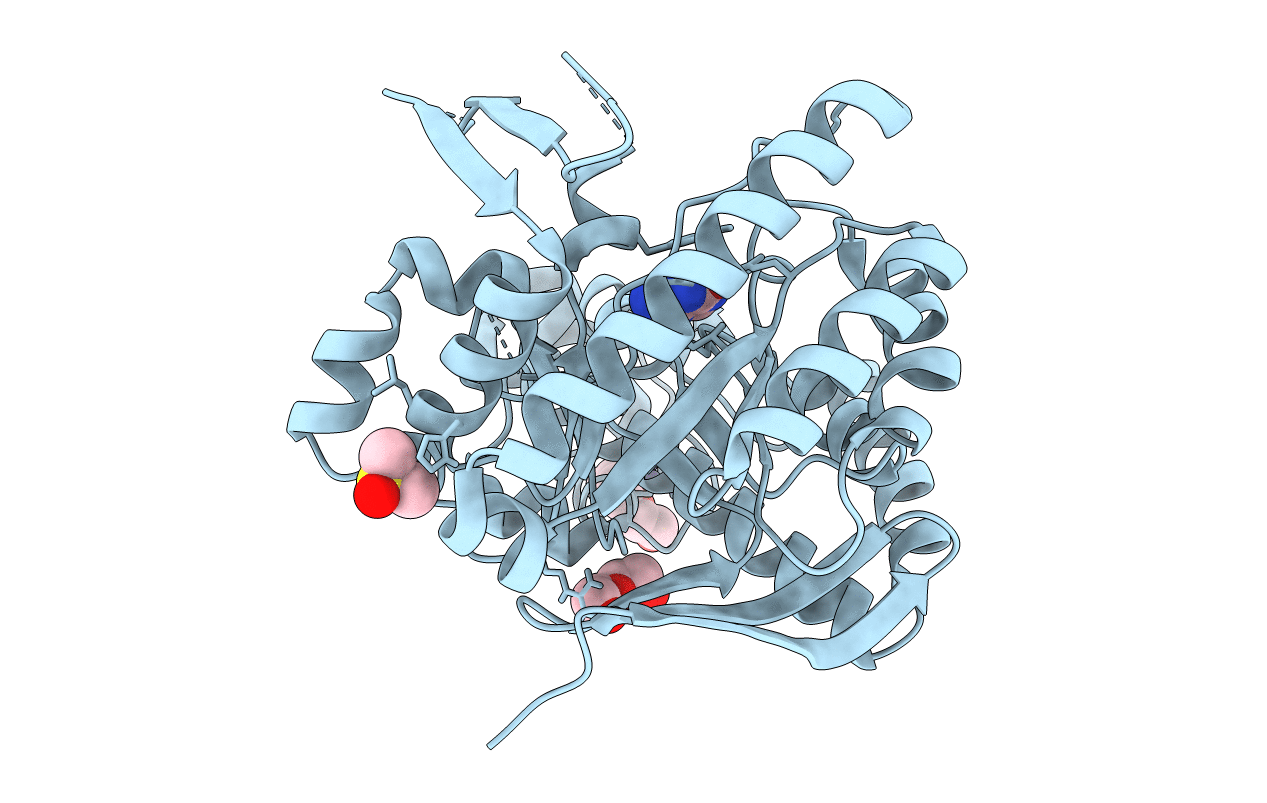
Crystal Structure Of Tgt In Complex With 4-(Aminomethane)Cyclohexane-1-Carboxylic Acid
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2018-05-16 Classification: TRANSFERASE Ligands: ZN, AMH, PEG, DMS, PGE |
 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2018-03-07 Classification: TRANSFERASE Ligands: ZN, DMS, PEG, GUN, PGE |
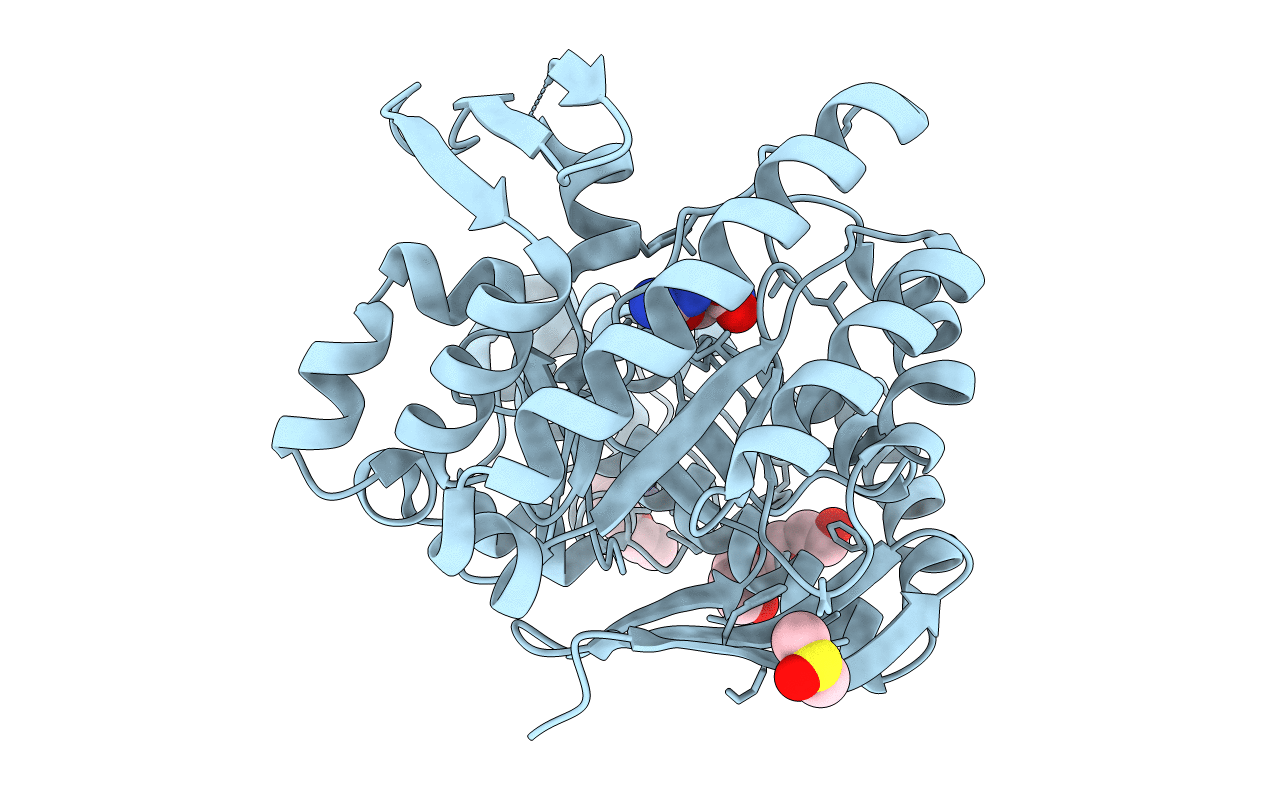 |
Organism: Zymomonas mobilis subsp. mobilis zm4 = atcc 31821
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2018-03-07 Classification: TRANSFERASE Ligands: ZN, DMS, GGB, PGE, PG4 |
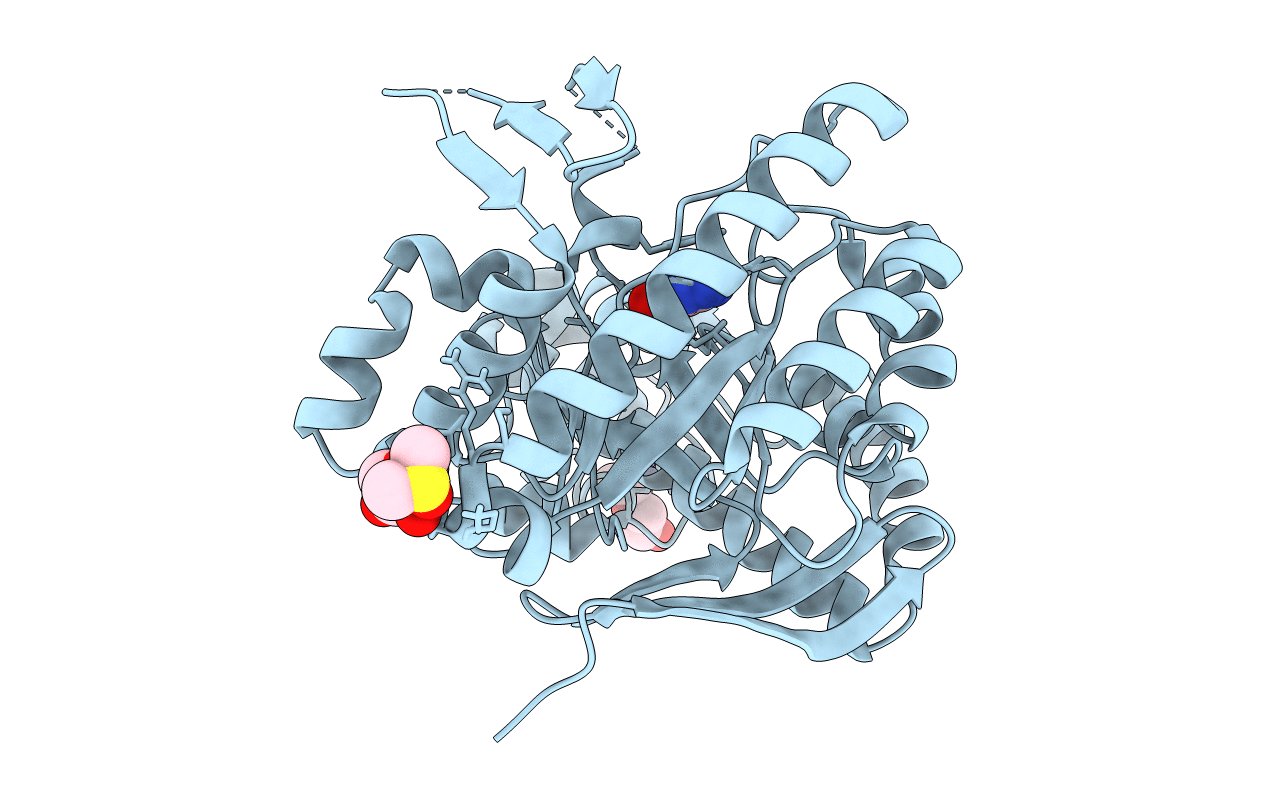 |
Crystal Structure Of Tgt In Complex With 2,6-Dioxy-8-Azapurine, 2,6-Dioxy-8-Azapurine, 2,6-Dioxy-8-Azapurine
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-03-07 Classification: TRANSFERASE Ligands: ZN, PEG, DMS, AZA |
 |
Crystal Structure Of Tgt In Complex With 3-Pyridinecarboxylic Acid, 6-(Dimethylamino)
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2017-07-19 Classification: TRANSFERASE Ligands: ZN, DMS, 46L, PEG, PG4 |

