Search Count: 15
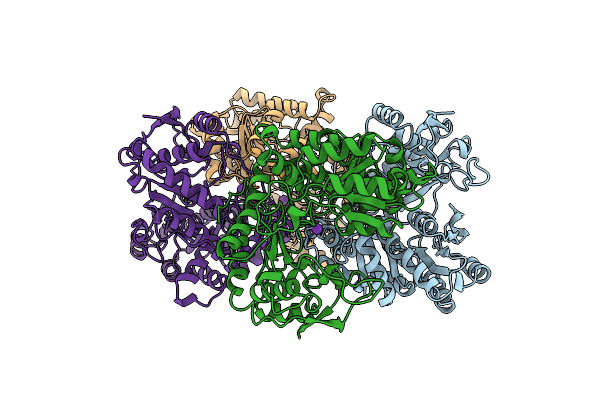 |
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1998-01-28 Classification: TRYPTOPHAN BIOSYNTHESIS Ligands: K |
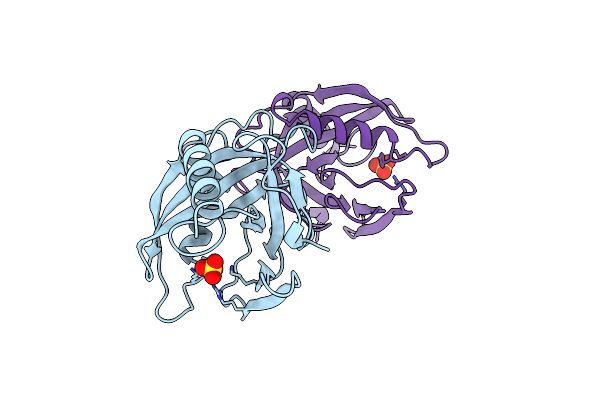 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 1997-12-03 Classification: HYDROLASE Ligands: SO4 |
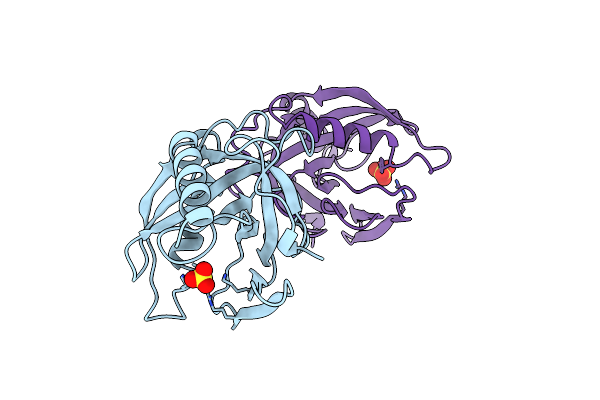 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 1997-12-03 Classification: HYDROLASE Ligands: SO4 |
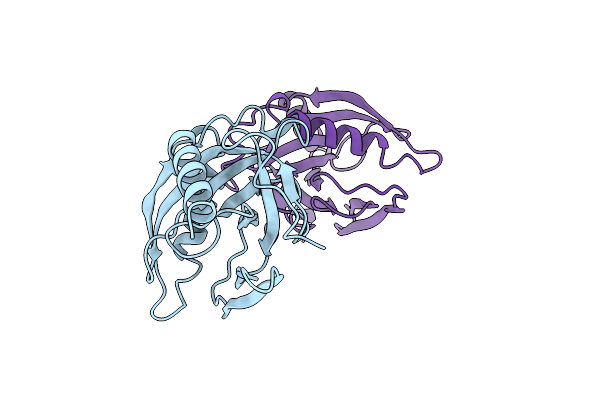 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-12-03 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1997-12-03 Classification: HYDROLASE |
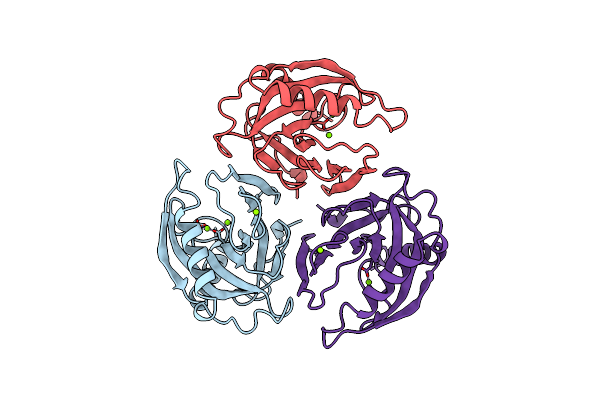 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1997-09-04 Classification: HYDROLASE Ligands: MG |
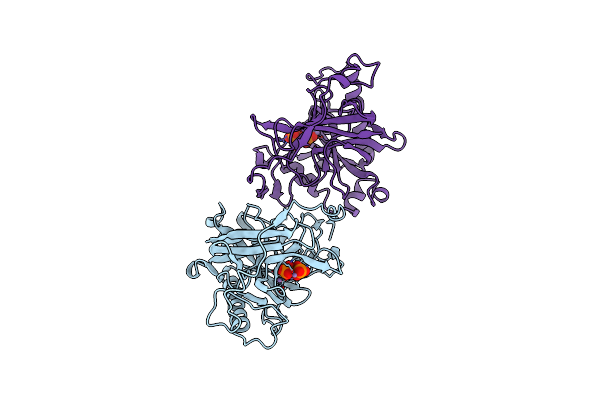 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1996-12-07 Classification: HYDROLASE Ligands: MN, PO4 |
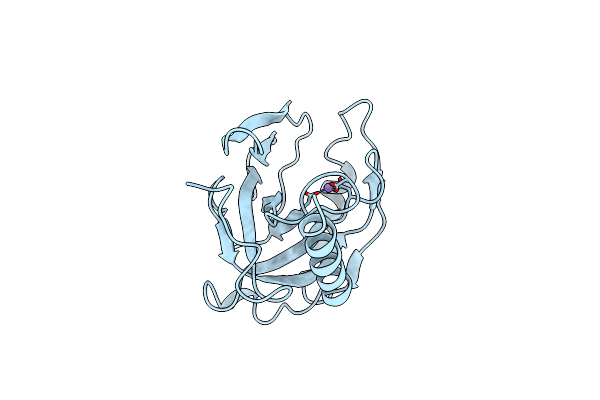 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1996-04-03 Classification: ACID ANHYDRIDE HYDROLASE Ligands: MN |
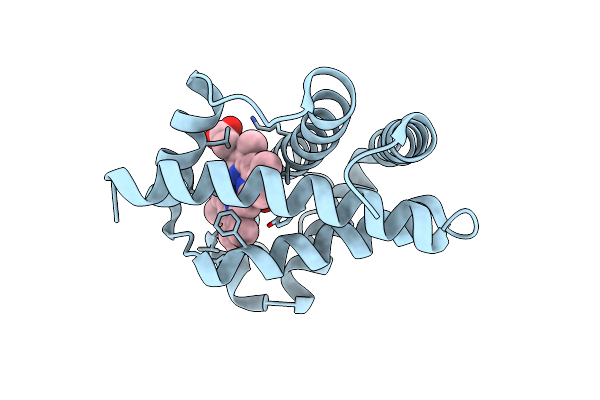 |
Crystal Structure Of Ferric Complexes Of The Yellow Lupin Leghemoglobin With Isoquinoline At 1.8 Angstroms Resolution (Russian)
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-02-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
 |
Crystal Structure Of Ferric Complexes Of The Yellow Lupin Leghemoglobin With Isoquinoline At 1.8 Angstroms Resolution (Russian)
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-02-27 Classification: OXYGEN TRANSPORT Ligands: HEM, NO |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-01-26 Classification: OXIDOREDUCTASE(ALDEHYDE(D),NAD+(A)) Ligands: SO4 |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 1995-01-26 Classification: OXIDOREDUCTASE(ALDEHYDE(D),NAD+(A)) Ligands: AZI, SO4, NAD |
 |
X-Ray Crystallographic Studies Of Recombinant Inorganic Pyrophosphatase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-12-20 Classification: ACID ANHYDRIDE HYDROLASE |
 |
Organism: Thermoactinomyces vulgaris
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 1994-01-31 Classification: HYDROLASE(SERINE PROTEASE) Ligands: CA, NA, SO4 |
 |
Organism: Citrobacter intermedius
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1993-10-31 Classification: LYASE(CARBON-CARBON) Ligands: SO4 |

