Search Count: 21
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: CA, U7F, ZN |
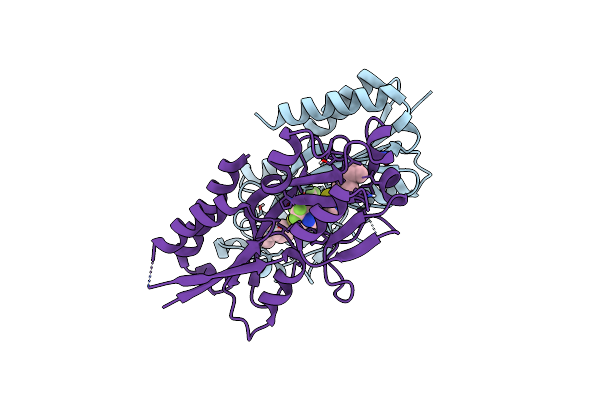 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Compound N-((2-(4-Cyclopropylphenyl)Thiazol-5-Yl)Methyl)-2-(Trifluoromethyl)Pyridin-4-Amine
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-11-30 Classification: GENE REGULATION Ligands: XBH |
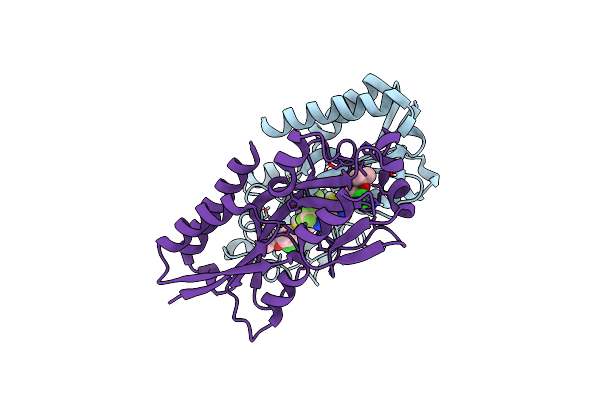 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Compound 1456
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-11-23 Classification: GENE REGULATION Ligands: 9ZL |
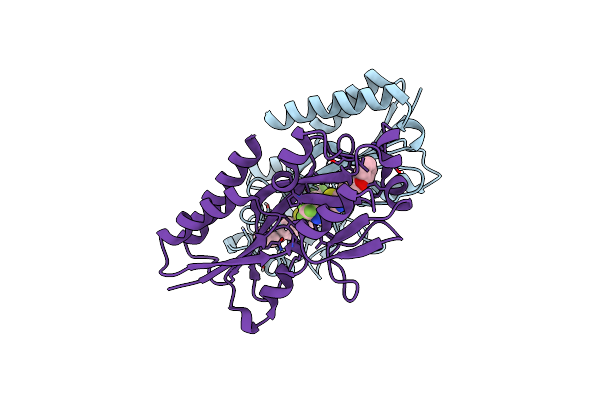 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Compound N-((2-(4-Phenoxyphenyl)Thiazol-5-Yl)Methyl)-2-(Trifluoromethyl)Pyridin-4-Amine
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-11-23 Classification: GENE REGULATION Ligands: A0F |
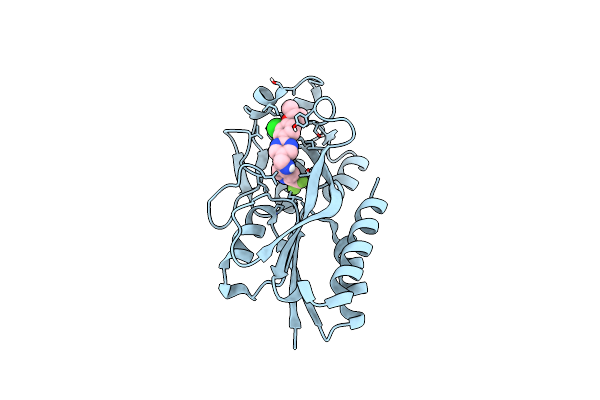 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With 3-Pyridin-4-Yl-2,4-Dihydro-Indeno[1,2-.C.]Pyrazole
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-07-27 Classification: GENE REGULATION Ligands: 5N9 |
 |
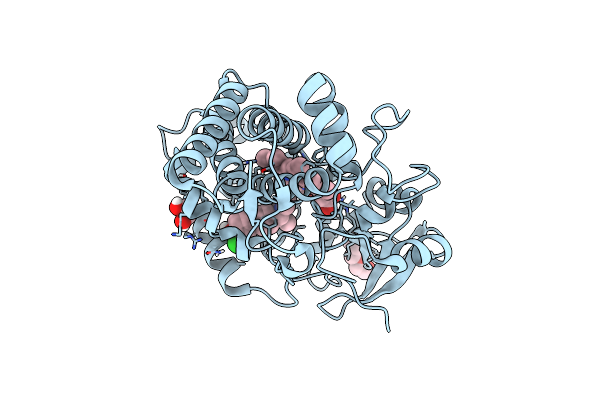
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-19 Classification: HYDROLASE Ligands: ZN, V85, CA |
 |
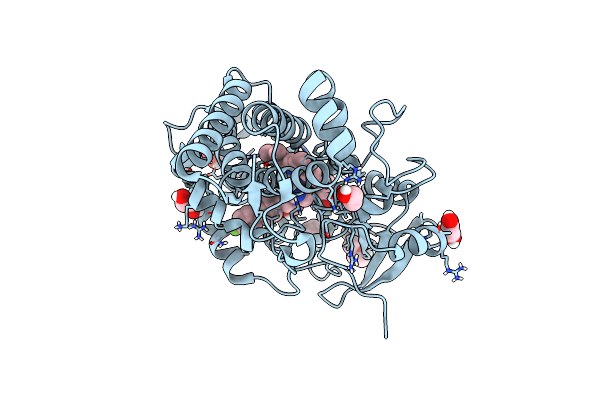
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: N55, PGE, EDO, HEM |
 |
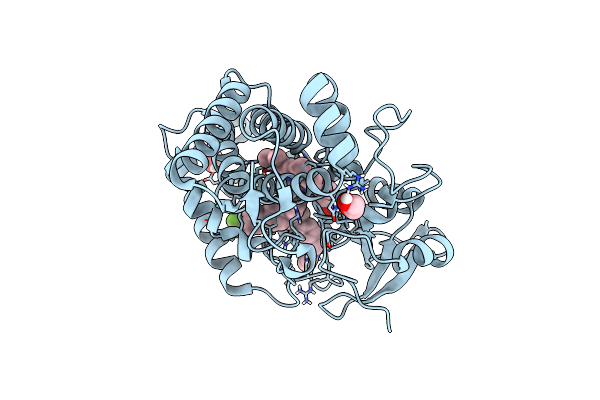
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: EDO, N5Z, PGE, HEM |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: N5W, EDO, HEM |
 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Triazolo-Pyridine Inverse Agonist A
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2021-04-14 Classification: DNA BINDING PROTEIN Ligands: OT2, OT8, MG |
 |
Escherichia Coli Rna Polymerase And Ureidothiophene-2-Carboxylic Acid Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.02 Å Release Date: 2020-10-07 Classification: TRANSCRIPTION Ligands: QZY, ZN, MG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-05 Classification: HYDROLASE Ligands: EEK, ZN, CA, SO4, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-03-28 Classification: HYDROLASE Ligands: CA, ZN, CXH |
 |
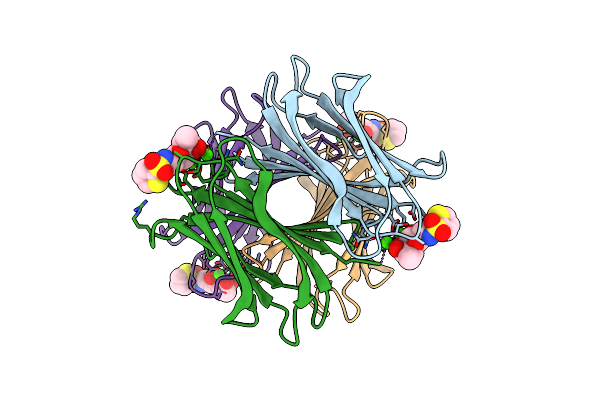
Crystal Structure Of The Peptidase Domain Of Collagenase H From Clostridium Histolyticum In Complex With N-Aryl Mercaptoacetamide-Based Inhibitor
Organism: Hathewaya histolytica
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: ZN, CA, 9NB |
 |
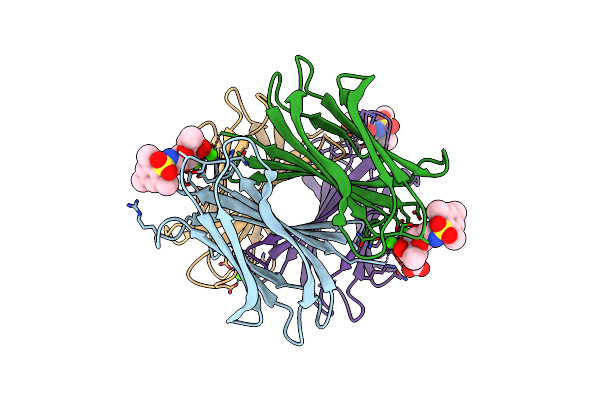
Structure Of The Lecb Lectin From Pseudomonas Aeruginosa Strain Pa14 In Complex With 2-Thiophenesulfonamide-N-(Beta-L-Fucopyranosyl Methyl)
Organism: Pseudomonas aeruginosa (strain ucbpp-pa14)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-12-20 Classification: SUGAR BINDING PROTEIN Ligands: CA, FUL, PK6 |
 |
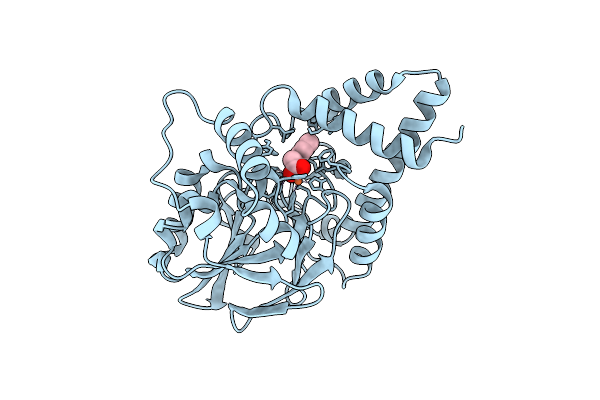
Structure Of The Lecb Lectin From Pseudomonas Aeruginosa Strain Pa14 In Complex With 2,4,6-Trimethylphenylsulfonamide-N-Methyl-L-Fucopyranoside
Organism: Pseudomonas aeruginosa (strain ucbpp-pa14)
Method: X-RAY DIFFRACTION Release Date: 2017-12-20 Classification: SUGAR BINDING PROTEIN Ligands: CA, FUL, 7KT, FLC |
 |
Crystal Structure Of Pqs Response Protein Pqse In Complex With 2-Aminobenzoylacetate
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-04-27 Classification: METAL BINDING PROTEIN Ligands: FE, 61M |
 |
Crystal Structure Of Pqs Response Protein Pqse In Complex With 2-(Pyridin-3-Yl)Benzoic Acid
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2016-04-27 Classification: METAL BINDING PROTEIN Ligands: FE, 61O |
 |
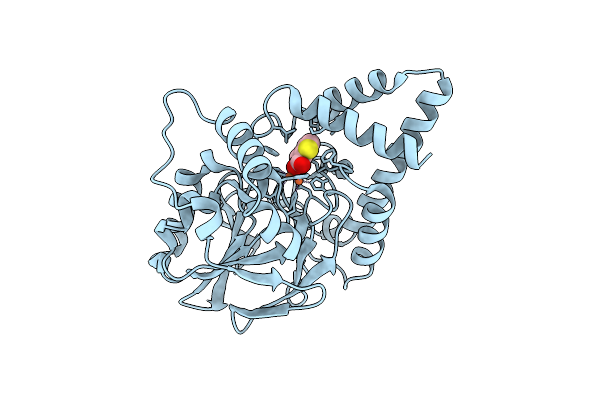
Crystal Structure Of Pqs Response Protein Pqse In Complex With 2-(1H-Pyrrol-1-Yl)Benzoic Acid
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-04-27 Classification: METAL BINDING PROTEIN Ligands: FE, 60Q |
 |
Crystal Structure Of Pqs Response Protein Pqse In Complex With 3-Methylthiophene-2-Carboxylic Acid
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-04-27 Classification: METAL BINDING PROTEIN Ligands: FE, 60P |

