Search Count: 95
 |
Cryoem Structure Of A C7-Symmetrical Groel7-Groes7 Cage In Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Disordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Ordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Structure Average Of Groel14 Complexes Found In The Cytosol Of Escherichia Coli Overexpressing Groel Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
In Situ Structure Average Of Groel14-Groes14 Complexes In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Cryoem Structure Of A Groel14-Groes7 Complex In Presence Of Adp-Befx With Wide Groel7 Trans Ring Conformation
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel14-Groes7 Complex In Presence Of Adp-Befx With Narrow Groel7 Trans Ring Conformation
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
In Situ Structure Average Of Groel14-Groes7 Complexes With Wide Groel7 Trans Ring Conformation In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
In Situ Structure Average Of Groel14-Groes7 Complexes With Narrow Groel7 Trans Ring Conformation In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-10-11 Classification: CHAPERONE Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-10-11 Classification: CHAPERONE Ligands: NAG |
 |
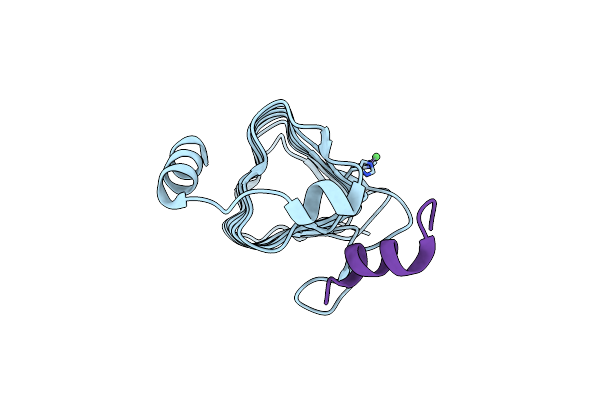
Crystal Structure Of The Carbonic Anhydrase-Like Domain Of Ccmm From Synechococcus Elongatus (Strain Pcc 7942)
Organism: Synechococcus elongatus (strain pcc 7942 / fachb-805)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-11-10 Classification: PHOTOSYNTHESIS Ligands: NI, CL |
 |
Crystal Structure Of The Carbonic Anhydrase-Like Domain Of Ccmm In Complex With The C-Terminal 17 Residues Of Ccaa From Synechococcus Elongatus (Strain Pcc 7942)
Organism: Synechococcus elongatus (strain pcc 7942 / fachb-805)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-11-10 Classification: PHOTOSYNTHESIS Ligands: NI, CL |
 |
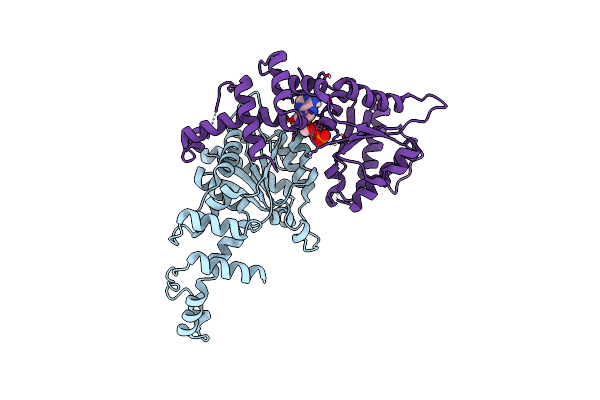
Crystal Structure Of The Aaa Domain Of Rubisco Activase From Nostoc Sp. (Strain Pcc 7120), Gadolinium Complex
Organism: Nostoc sp. pcc 7120 = fachb-418
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2020-09-23 Classification: CHAPERONE Ligands: GD, CL, ADP |
 |
Crystal Structure Of The Aaa Domain Of Rubisco Activase From Nostoc Sp. (Strain Pcc 7120)
Organism: Nostoc sp. pcc 7120 = fachb-418
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-09-23 Classification: CHAPERONE Ligands: CL, ADP |
 |
Cryoem Structure Of Rubisco Activase With Its Substrate Rubisco From Nostoc Sp. (Strain Pcc7120)
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: ELECTRON MICROSCOPY Release Date: 2020-09-23 Classification: CHAPERONE Ligands: ADP, AGS, MG, CAP |
 |
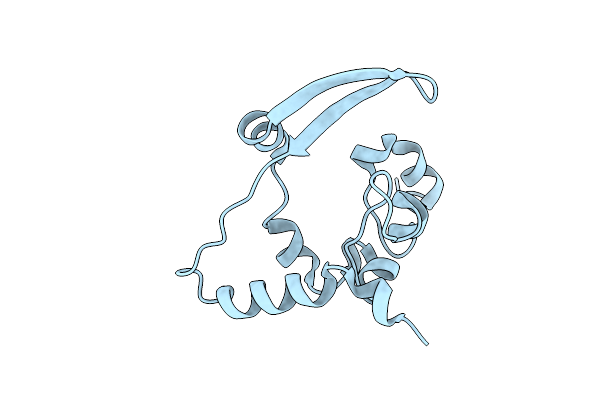
Cryoem Structure Of The Interaction Between Rubisco Activase Small-Subunit-Like (Ssul) Domain With Rubisco From Nostoc Sp. (Strain Pcc7120)
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: ELECTRON MICROSCOPY Release Date: 2020-09-23 Classification: PHOTOSYNTHESIS |
 |
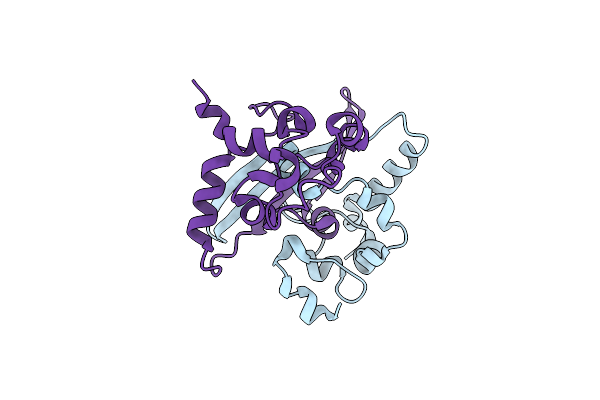
Crystal Structure Of C-Terminal Dimerization Domain Of Nucleocapsid Phosphoprotein From Sars-Cov-2, Crystal Form Ii
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN |
 |
1.45 Angstrom Resolution Crystal Structure Of C-Terminal Dimerization Domain Of Nucleocapsid Phosphoprotein From Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-05-20 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Chaperonin Gp146 From The Bacteriophage El 2 (Pseudomonas Aeruginosa) In Presence Of Atp-Befx, Crystal Form I
Organism: Pseudomonas phage el
Method: X-RAY DIFFRACTION Resolution:4.03 Å Release Date: 2020-04-22 Classification: CHAPERONE Ligands: MG, ATP |

