Search Count: 54
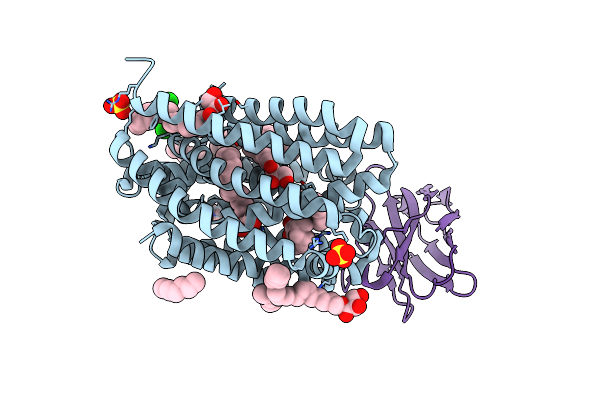 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 2.7552 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, OCT, SO4, ZN, CL |
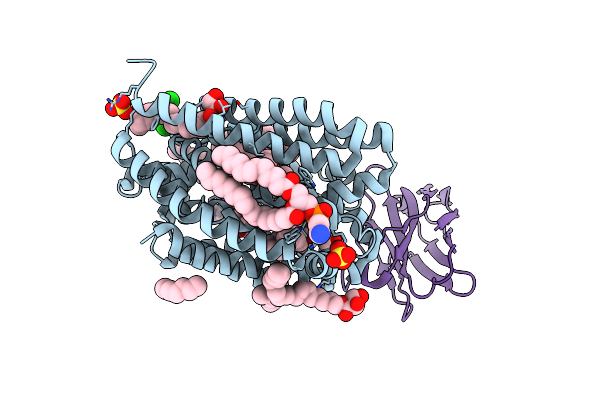 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 0.9688 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, LOP, OCT, CL, SO4, ZN |
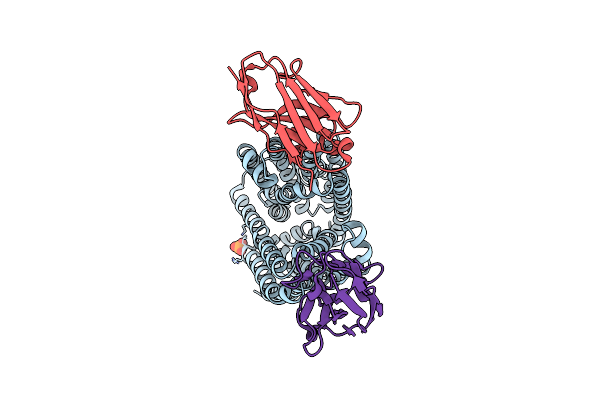 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: PO4 |
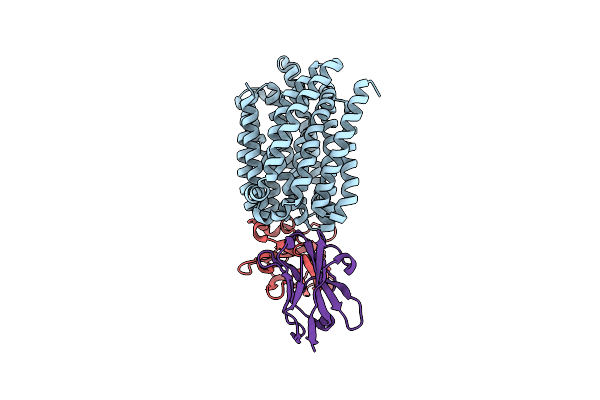 |
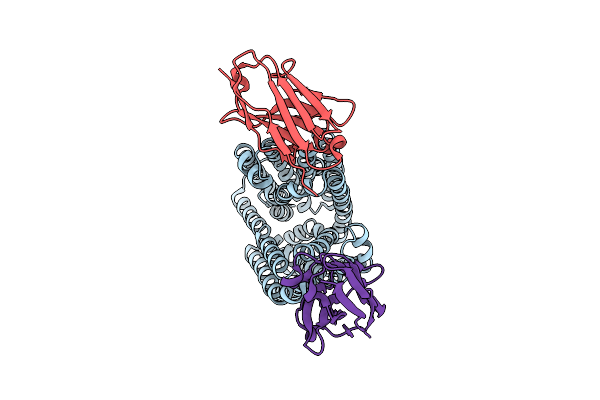
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
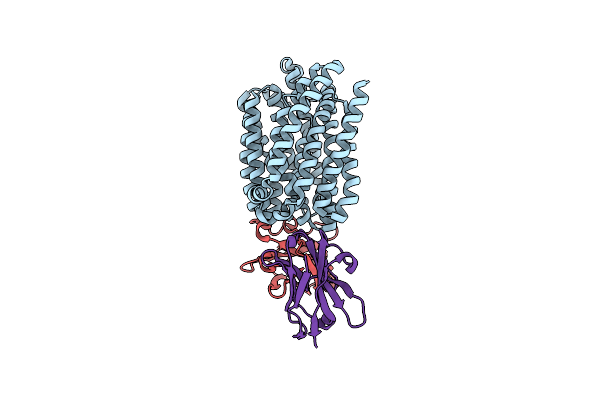
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
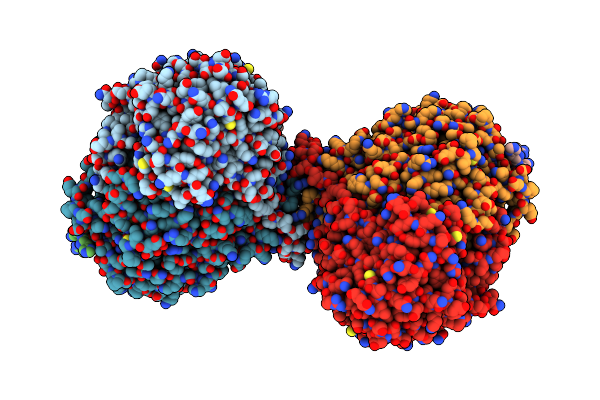 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae s288c , Saccharomyces cerevisiae, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR Ligands: PLP, Z1T |
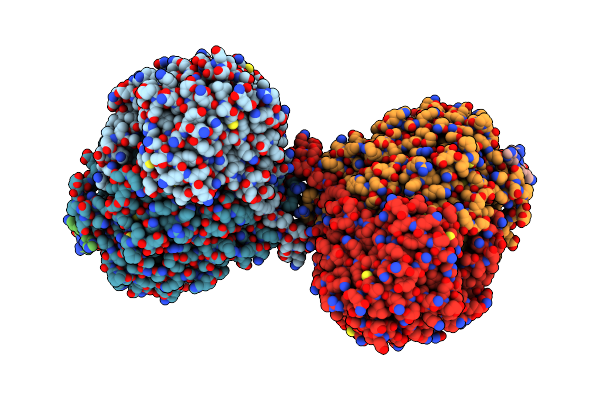 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR |
 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR Ligands: PLP, Z1T |
 |
Native Thermogutta Terrifontis Endoglucanase Catalytic Domain With A Linker At C-Terminal From Glycoside Hydrolase Family 5 (Ttend5A-Cdc)
Organism: Thermogutta terrifontis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-10-12 Classification: HYDROLASE Ligands: SO4 |
 |
Thermogutta Terrifontis Endoglucanase Of Glycoside Hydrolase Family 5 (Ttend5A)
Organism: Thermogutta terrifontis
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-08-17 Classification: HYDROLASE Ligands: SO4 |
 |
E329A Mutant Thermogutta Terrifontis Endoglucanase Catalytic Domain With C-Linker From Glycoside Hydrolase Family 5 (Ttend5A-Cdc-E329A)
Organism: Thermogutta terrifontis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-08-17 Classification: HYDROLASE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Escherichia virus t4, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Escherichia virus t4, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Escherichia virus t4, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike : H11-H4 Q98R H100E Nanobody Complex In 1Up2Down Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Escherichia virus t4, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Tequatrovirus t4, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2022-07-13 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike : H11-H4 Q98R H100E Nanobody Complex In 2Up1Down Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Escherichia virus t4, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
1.58 A Structure Of Human Apoferritin Obtained From Titan Krios 2 At Ebic, Dls Under Commissioning Session Cm26464-2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:1.60 Å Release Date: 2022-06-22 Classification: METAL BINDING PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: VIRAL PROTEIN Ligands: NAG, EIC |

