Search Count: 51
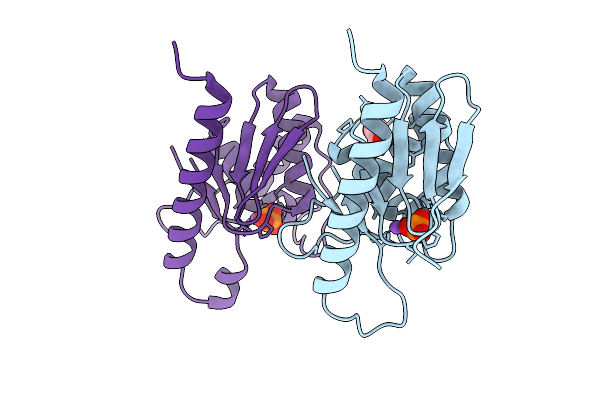 |
High Resolution Structure Of Cytidine Deaminase T6S Toxin From Pseudomonas Syringae
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TOXIN Ligands: ZN, PO4, EDO, NA |
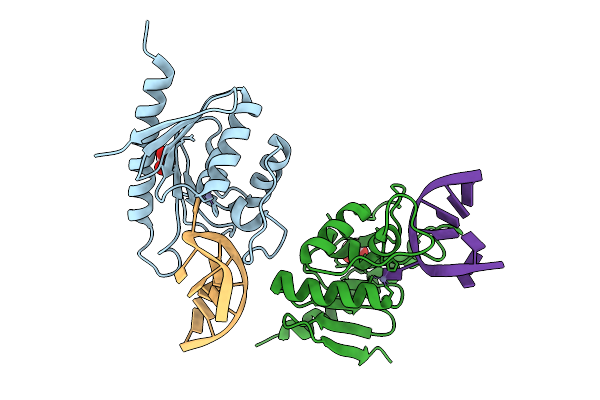 |
Cytidine Deaminase T6S Toxin From Pseudomonas Syringae Complexed With Substrate Dna
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TOXIN/DNA Ligands: ZN, EDO |
 |
Crystal Structure Of Sars-Cov-2 Omicron Main Protease (Mpro) Complex With Azapeptide Inhibitor 20A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: A1BKV |
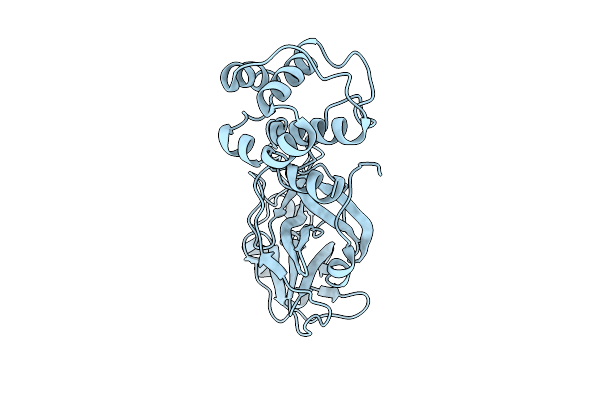 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE |
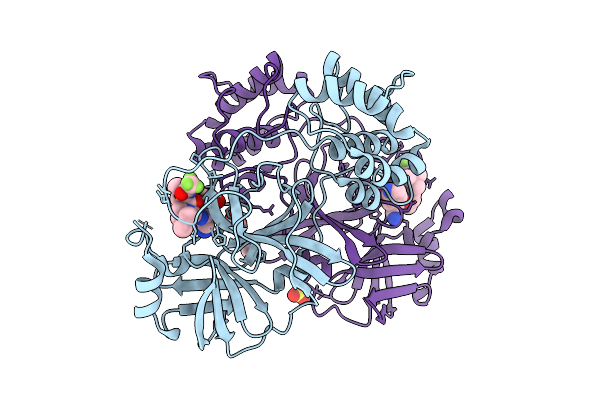 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Quadruple Mutants In Complex With Nirmatrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: 4WI, EDO, BR, SO4 |
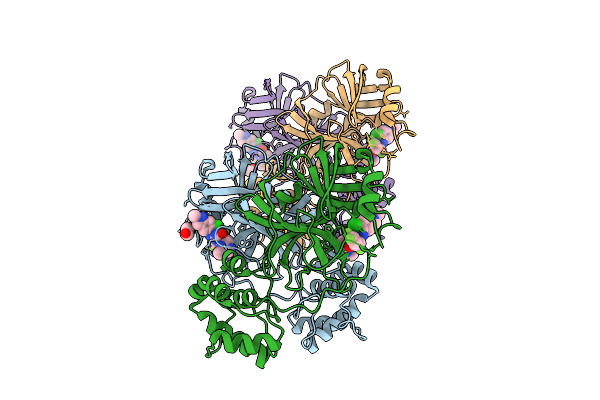 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Quadruple Mutants In Complex With Ensitrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: 7YY, EDO |
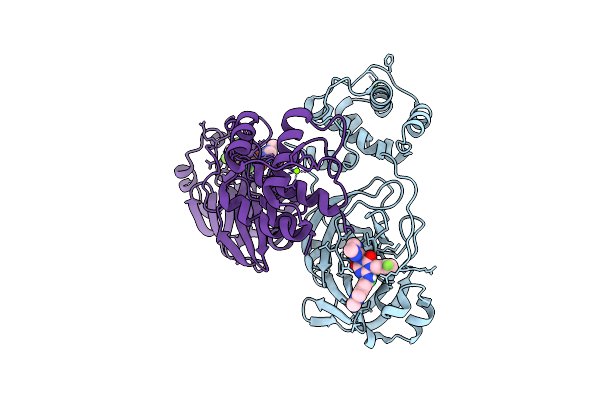 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Double Mutants In Complex With Ensitrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: 7YY, MG, CL |
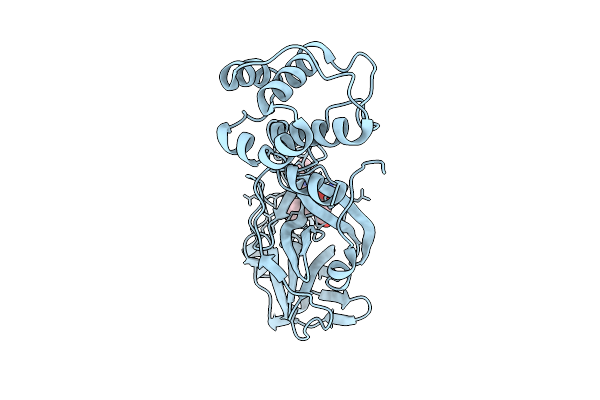 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Triple Mutants In Complex With Bofutrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: FHR |
 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Quadruple Mutants In Complex With Bofutrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: FHR |
 |
Structure Of Sars-Cov-2 Main Protease In Complex With Bofutrelvir In Orthorhombic Form
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: FHR, EDO, PEG |
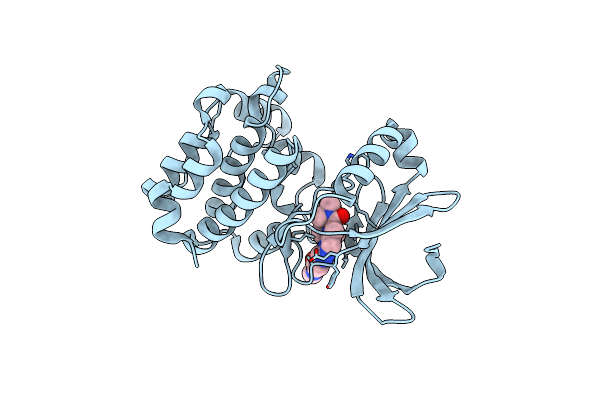 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: SIGNALING PROTEIN Ligands: OG0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-01-15 Classification: SIGNALING PROTEIN Ligands: OGR |
 |
Crystal Structure Of A Stem-Loop Dna Aptamer Complexed With Sars-Cov-2 Nucleocapsid Protein Rna-Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-11-13 Classification: VIRAL PROTEIN Ligands: EDO |
 |
Wild Type Apobec3A In Complex With Tt(Fdz)-Hairpin Inhibitor (Crystal Form 1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: PO4, ZN |
 |
Wild Type Apobec3A In Complex With Tt(Fdz)-Hairpin Inhibitor (Crystal Form 2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: ZN, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, CL, PO4, IHP |
 |
Zinc-Free Apobec3A (Inactive E72A Mutant) In Complex With Ttc-Hairpin Dna Substrate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: IHP, CL |
 |
Structure Of Apobec3A (E72A Inactive Mutant) In Complex With Ttc-Hairpin Dna Substrate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: IHP, ZN, CL, PO4 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-07-26 Classification: HYDROLASE/INHIBITOR Ligands: O5F |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-07-26 Classification: HYDROLASE/INHIBITOR Ligands: O5O |

