Search Count: 22
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-04 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP, MG, ACY, GOL, PO4 |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, PO4, MG, ACY, GOL, PEG, NA |
 |
F420-2/Gtp(Gdp) Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-02-28 Classification: LIGASE Ligands: F42, GTP, GDP, MN, NA, SO4 |
 |
Nmr Shows Why A Small Chemical Change Almost Abolishes The Antimicrobial Activity Of Gccf
Organism: Lactiplantibacillus plantarum
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
Gtp Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-04-12 Classification: LIGASE Ligands: CO3, MN, NA, GTP, SO4 |
 |
F420-1/Gdp Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-04-12 Classification: LIGASE Ligands: GDP, F4I, MN, NA |
 |
L-Glutamate/Gtp Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-04-12 Classification: LIGASE Ligands: GGL, GTP, MN |
 |
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.83 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC Ligands: TFA |
 |
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC Ligands: MOH |
 |
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.77 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC Ligands: HOH |
 |
Organism: Xylaria
Method: X-RAY DIFFRACTION Resolution:0.77 Å Release Date: 2019-09-11 Classification: ANTIBIOTIC |
 |
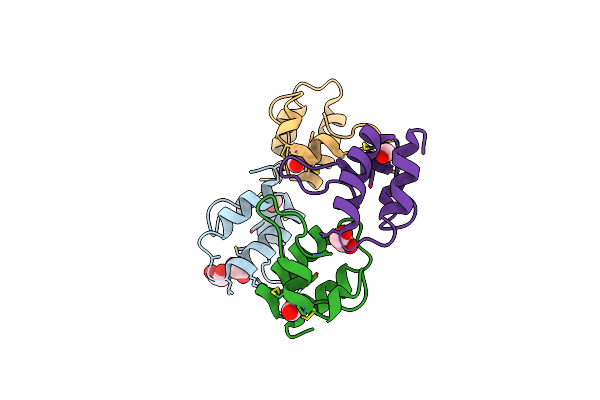
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-05-18 Classification: ANTIMICROBIAL PROTEIN |
 |
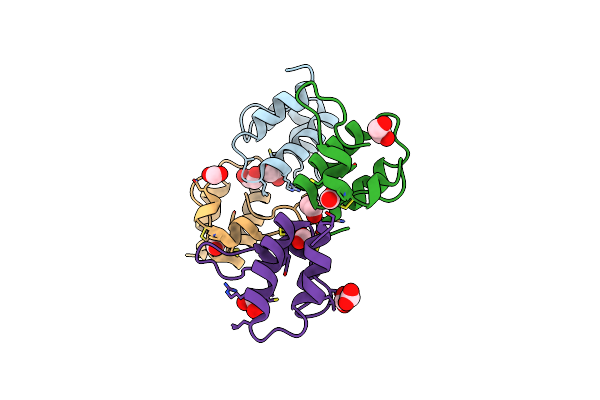
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2016-05-18 Classification: ANTIMICROBIAL PROTEIN Ligands: FMT, EDO |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2016-05-18 Classification: ANTIMICROBIAL PROTEIN Ligands: EDO, FMT |
 |
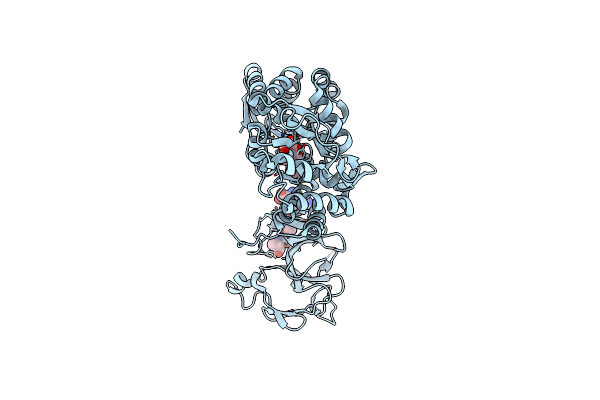
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-09-23 Classification: HYDROLASE |
 |
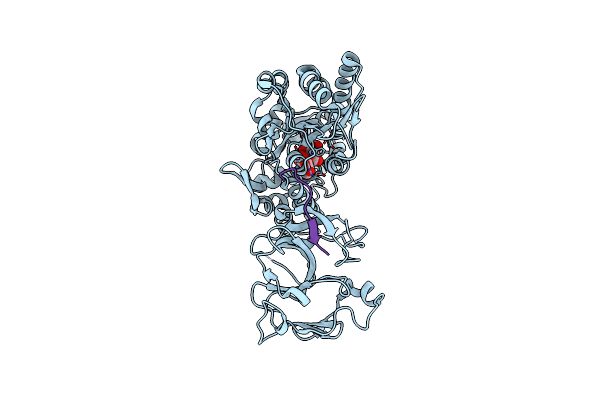
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb) In Complex With A Substrate Peptide (Pep1).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-09-23 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb) In Complex With A Substrate Peptide (Pep2).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-09-23 Classification: HYDROLASE |
 |
Crystal Structure Of The Type-I Signal Peptidase From Staphylococcus Aureus (Spsb) In Complex With An Inhibitor Peptide (Pep3).
Organism: Escherichia coli k-12, Staphylococcus aureus subsp. aureus str. newman, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-09-23 Classification: hydrolase/hydrolase inhibitor |
 |
Crystal Structure Of The Clr:Ramp2 Extracellular Domain Heterodimer With Bound Adrenomedullin
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-05-20 Classification: MEMBRANE PROTEIN/HORMONE Ligands: EDO |

