Search Count: 24
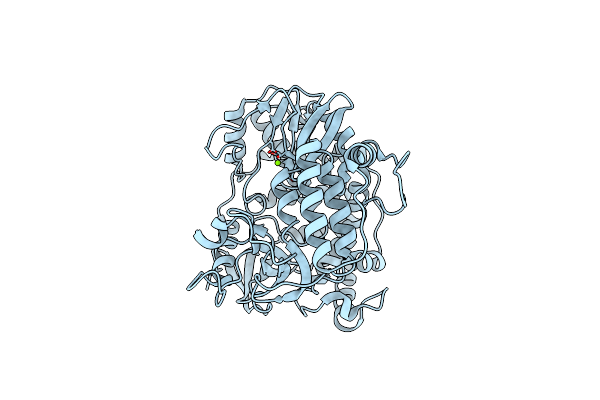 |
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: VIRAL PROTEIN Ligands: MG |
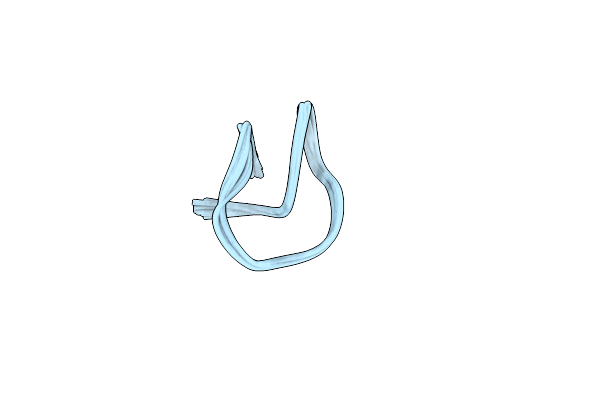 |
Organism: Lentzea jiangxiensis
Method: SOLUTION NMR Release Date: 2024-09-25 Classification: UNKNOWN FUNCTION |
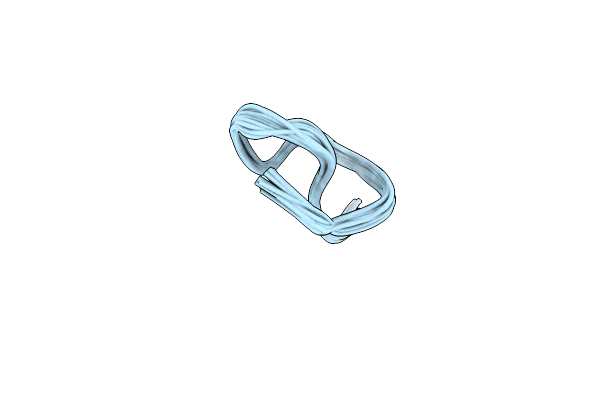 |
Organism: Streptomyces katrae
Method: SOLUTION NMR Release Date: 2024-09-25 Classification: UNKNOWN FUNCTION |
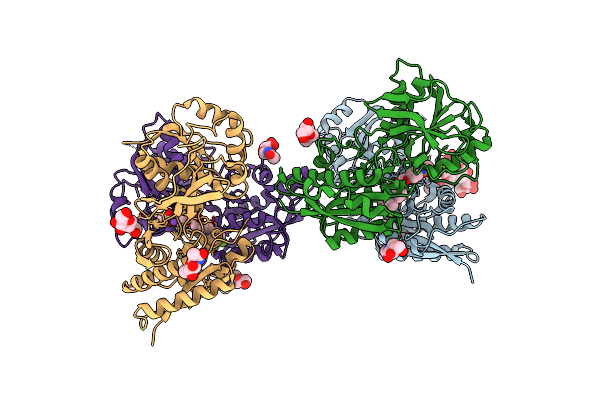 |
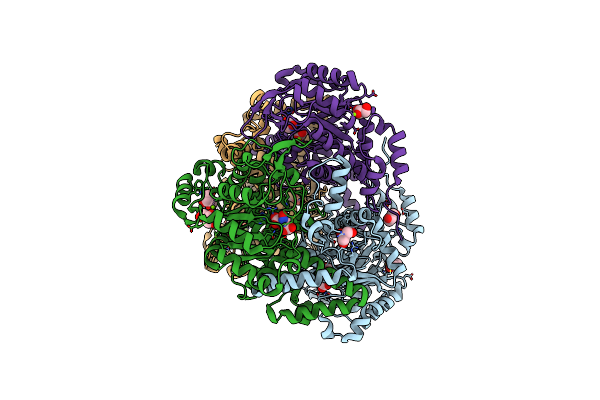
Heterodimer Of The Glun1B-Glun2B Nmda Receptor Amino-Terminal Domains Bound To Allosteric Inhibitor 93-108
Organism: Xenopus laevis, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-03-01 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: NAG, NA, YGW |
 |
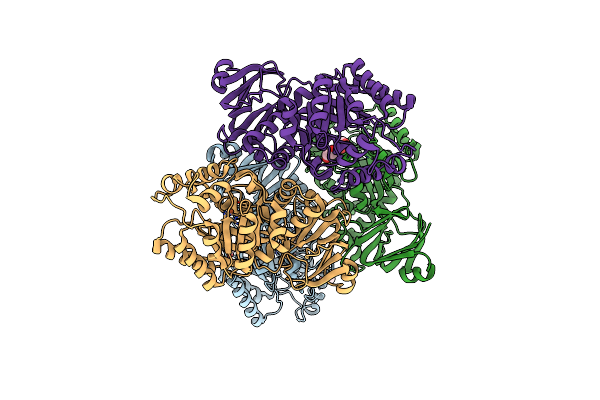
Isocitrate Lyase-1 From Mycobacterium Tuberculosis Covalently Modified By 5-Descarboxy-5-Nitro-D-Isocitric Acid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-12-01 Classification: LYASE Ligands: 48J, MG, 54I, GOL, GLV |
 |
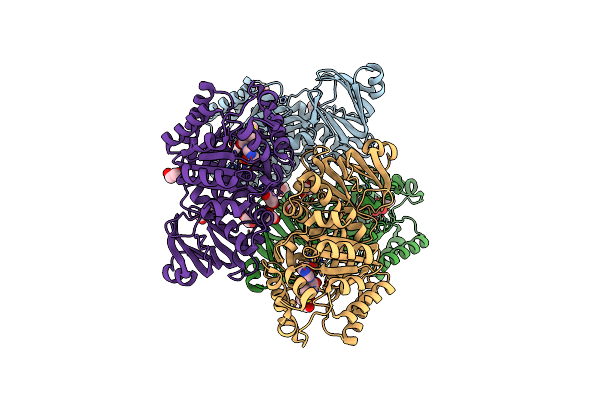
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: FE, EDO |
 |
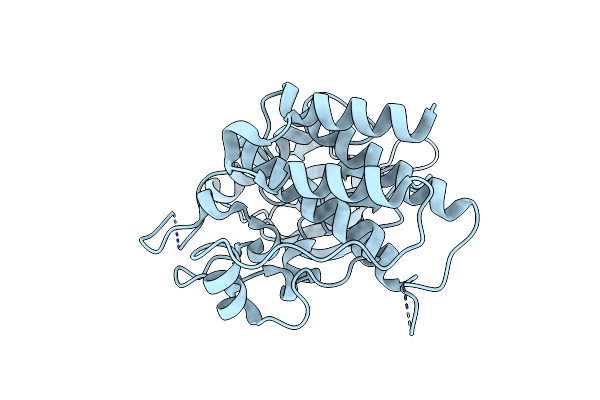
Crystal Structure Of Helicobacter Pylori Aminofutalosine Deaminase (Aflda) In Complex With Methylthio-Coformycin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-06-23 Classification: HYDROLASE/INHIBITOR Ligands: FE, EDO, MCF |
 |
Organism: Streptomyces avermitilis
Method: SOLUTION NMR Release Date: 2020-12-16 Classification: UNKNOWN FUNCTION |
 |
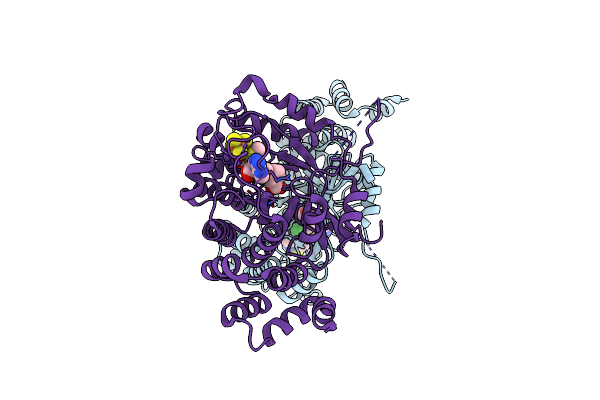
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-25 Classification: TRANSFERASE |
 |
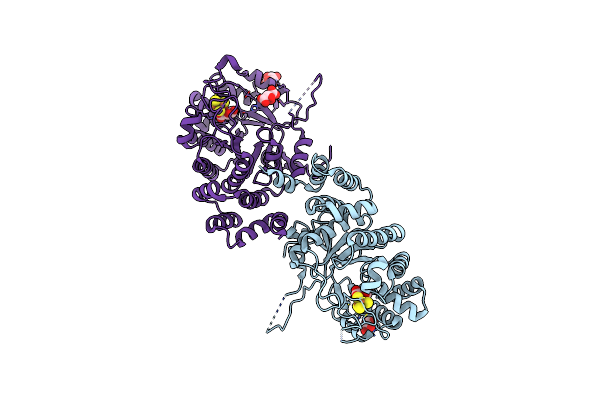
X-Ray Crystal Structure Of Mqne From Pedobacter Heparinus In Complex With Aminofutalosine And Methionine
Organism: Pedobacter heparinus (strain atcc 13125 / dsm 2366 / cip 104194 / jcm 7457 / nbrc 12017 / ncimb 9290 / nrrl b-14731 / him 762-3)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, MET, V47, CL |
 |
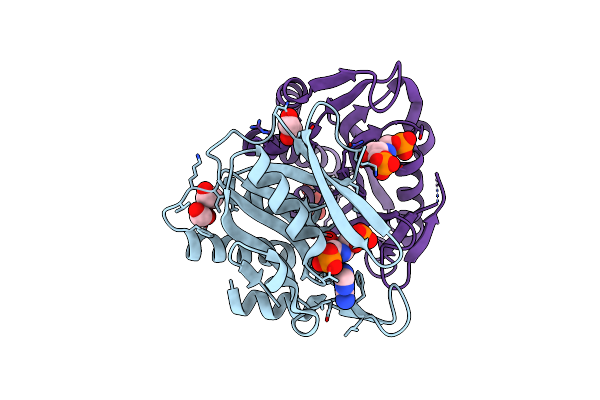
Organism: Pedobacter heparinus (strain atcc 13125 / dsm 2366 / cip 104194 / jcm 7457 / nbrc 12017 / ncimb 9290 / nrrl b-14731 / him 762-3)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, TAR |
 |
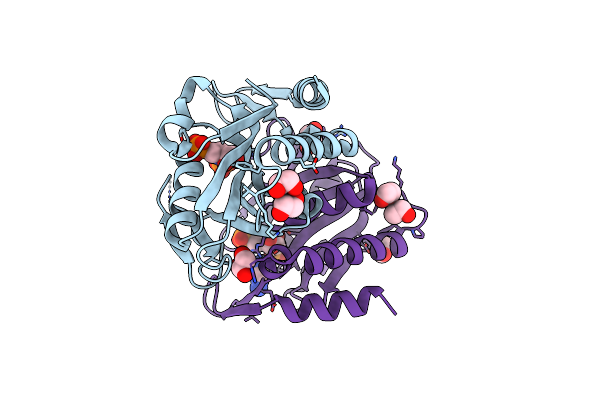
Crystal Structure Of Adenine Phosphoribosyltransferase From Saccharomyces Cerevisiae Complexed With D-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (D-Diab) And Adenine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-12-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: IR8, EDO, ADE |
 |
Crystal Structure Of Adenine Phosphoribosyltransferase From Saccharomyces Cerevisiae Complexed With L-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (L-Diab) And Adenine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-12-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: IR9, EDO, ADE |
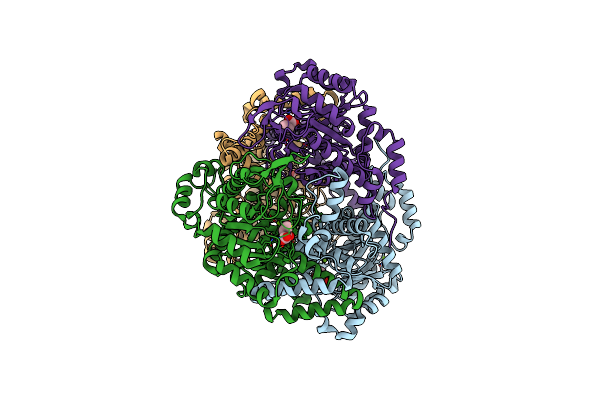 |
Crystal Structure Of 2-Vinyl Glyoxylate Modified Isocitrate Lyase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 35801 / tmc 107 / erdman)
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2016-09-14 Classification: LYASE/LYASE INHIBITOR Ligands: MG, VGX |
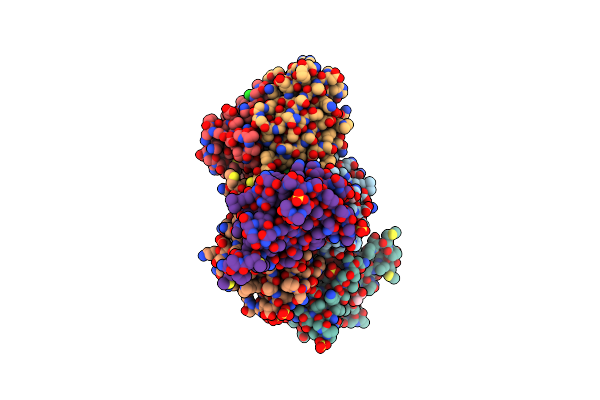 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-05-13 Classification: UNKNOWN FUNCTION Ligands: SO4, GOL, CL |
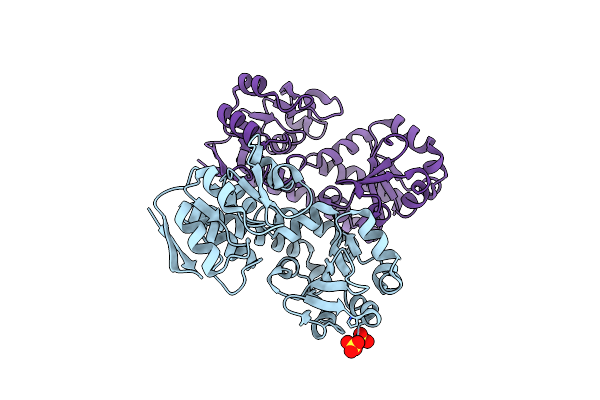 |
Structure Of A Putative Periplasmic Iron Siderophore Binding Protein (Rv0265C) From Mycobacterium Tuberculosis H37Rv
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-06-11 Classification: SOLUTE-BINDING PROTEIN Ligands: SO4, CL |
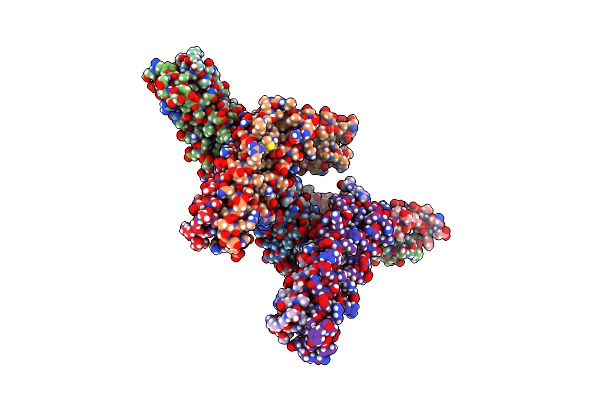 |
Crystal Structure Of The Mycobacterum Abscessus Esxef (Mab_3112-Mab_3113) Complex
Organism: Mycobacterium abscessus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2013-01-16 Classification: Structural Genomics, Unknown Function Ligands: SO4, BME, GOL |
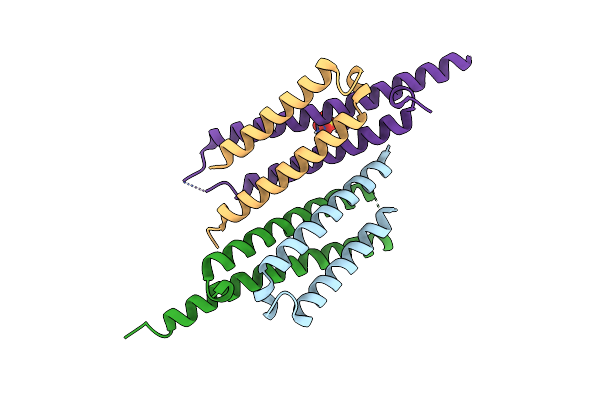 |
Crystal Structure Of The Mycobacterium Tuberculosis H37Rv Esxop (Rv2346C-Rv2347C) Complex In Space Group C2221
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2012-09-26 Classification: Structural Genomics, Unknown Function Ligands: SO4 |
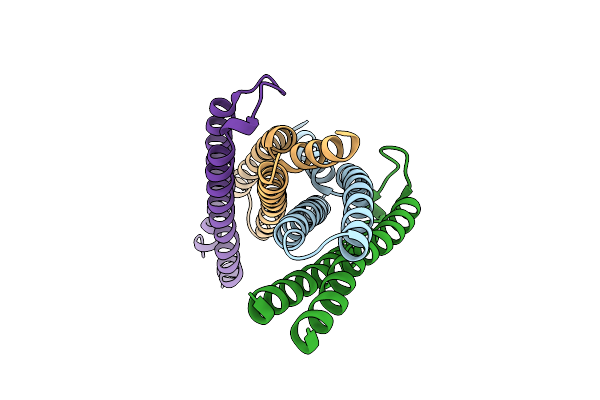 |
Crystal Structure Of The Mycobacterium Smegmatis Esxgh Complex (Msmeg_0620-Msmeg_0621)
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-01-26 Classification: METAL TRANSPORT |
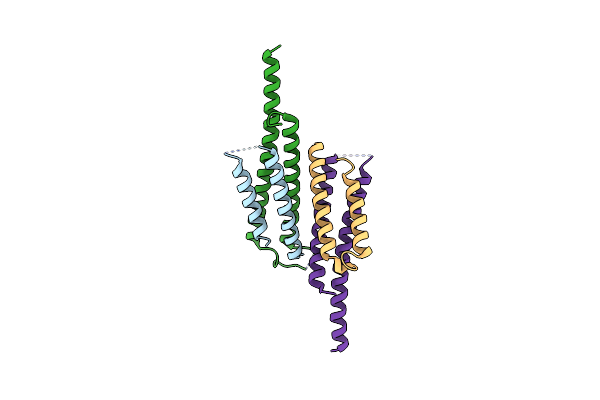 |
Crystal Structure Of The Mycobacterium Tuberculosis H37Rv Esxop Complex (Rv2346C-Rv2347C)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2010-08-25 Classification: Structural Genomics, Unknown function |

