Search Count: 45
 |
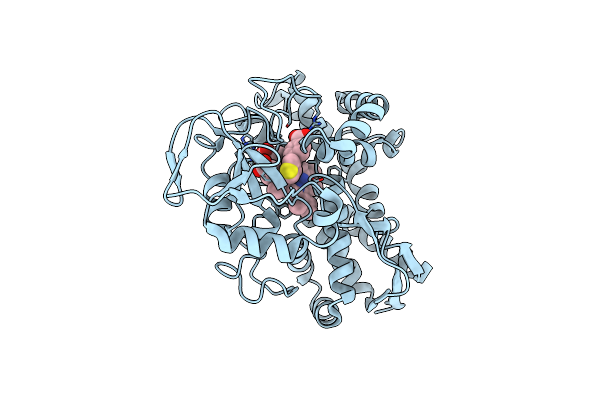
The Crystal Structure Of T252E Cyp199A4 Bound To 4-(Pyridin-2-Yl)Benzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: CL, HEM, PQS |
 |
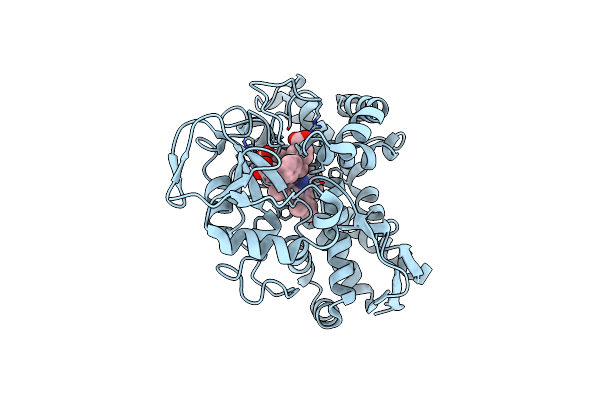
The Crystal Structure Of T252E Cyp199A4 Bound To 4-(Thiophen-3-Yl)Benzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: EJD, HEM, CL |
 |
The Crystal Structure Of The T252E Mutant Of Cyp199A4 Bound To 4-Benzylbenzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: IRJ, HEM, CL |
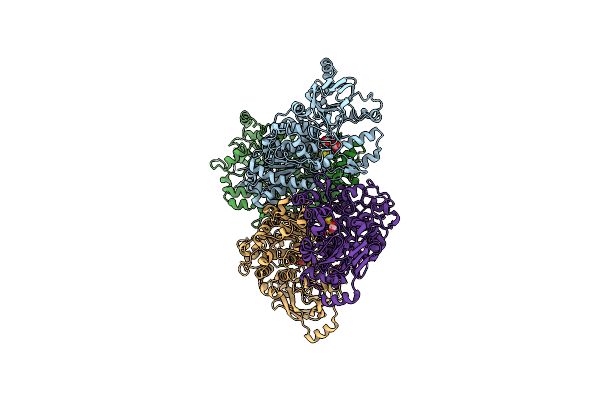 |
Crystal Structure Of Fe-S Cluster-Dependent Dehydratase From Paralcaligenes Ureilyticus In Complex With Mg
Organism: Paralcaligenes ureilyticus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-08-30 Classification: LYASE Ligands: FES, MG, BCT, CO2 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-05-10 Classification: TRANSCRIPTION Ligands: A8R |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-05-10 Classification: TRANSCRIPTION Ligands: A8R |
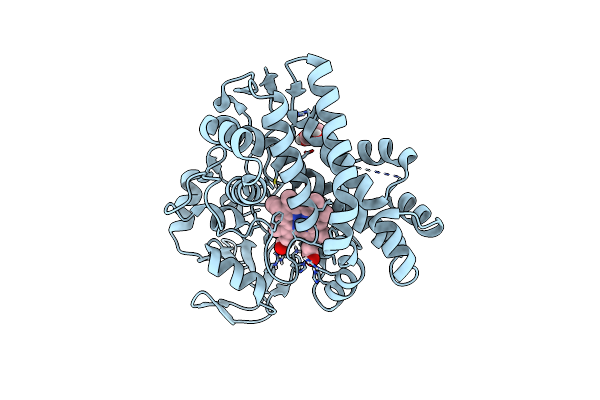 |
P450 (Oxya) From Kistamicin Biosynthesis, Mixed Heme Conformation, Attenuated Beam
Organism: Actinomadura parvosata subsp. kistnae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: GOL, HEM |
 |
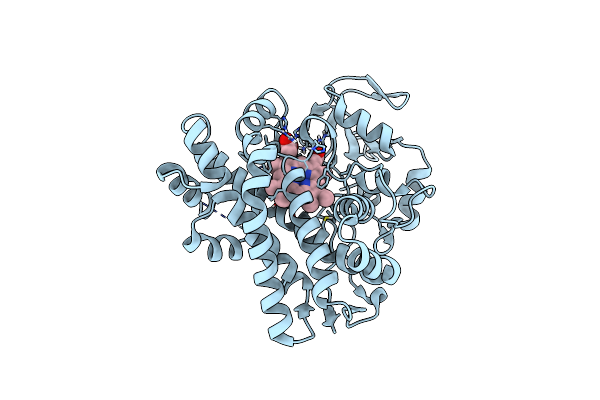
Organism: Actinomadura parvosata subsp. kistnae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
 |
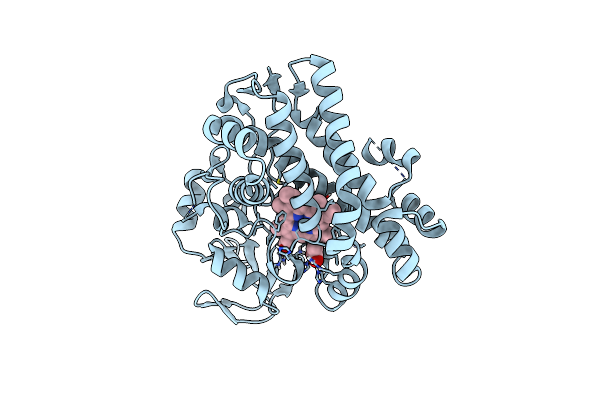
Organism: Actinomadura parvosata subsp. kistnae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
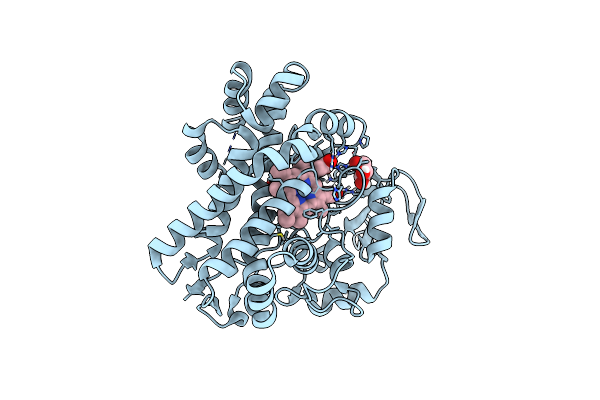
Organism: Actinomadura parvosata subsp. kistnae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
 |
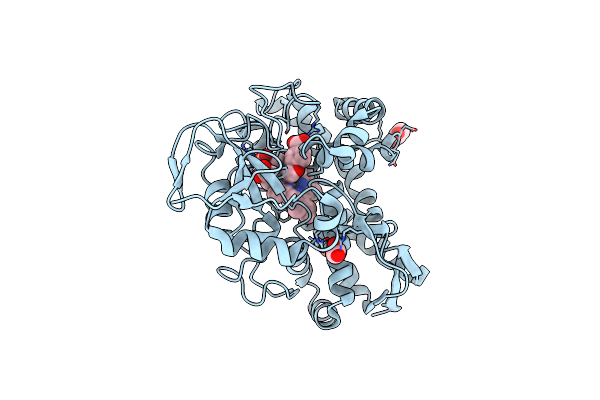
Organism: Actinomadura parvosata subsp. kistnae
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: IMD, HEM, GOL |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE Ligands: ANN, CL, MG, ACT, HEM |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: CL, HEM, MW7 |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: CL, HEM, MW7 |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: EPE, MGD, MO, CL, O |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: O, MGD, MO |
 |
Crystal Structure Of The Zn(Ii)-Bound Znua-Like Domain Of Streptococcus Pneumoniae Adca
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: ZN, CL |
 |
Crystal Structure Of The Zint-Like Domain Of Streptococcus Pneumoniae Adca In The Apo Form
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDT, NA |
 |
Crystal Structure Of Zn(Ii)-Bound Zint-Like Domain Of Streptococcus Pneumoniae Adca
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: ZN, NA, MG |

